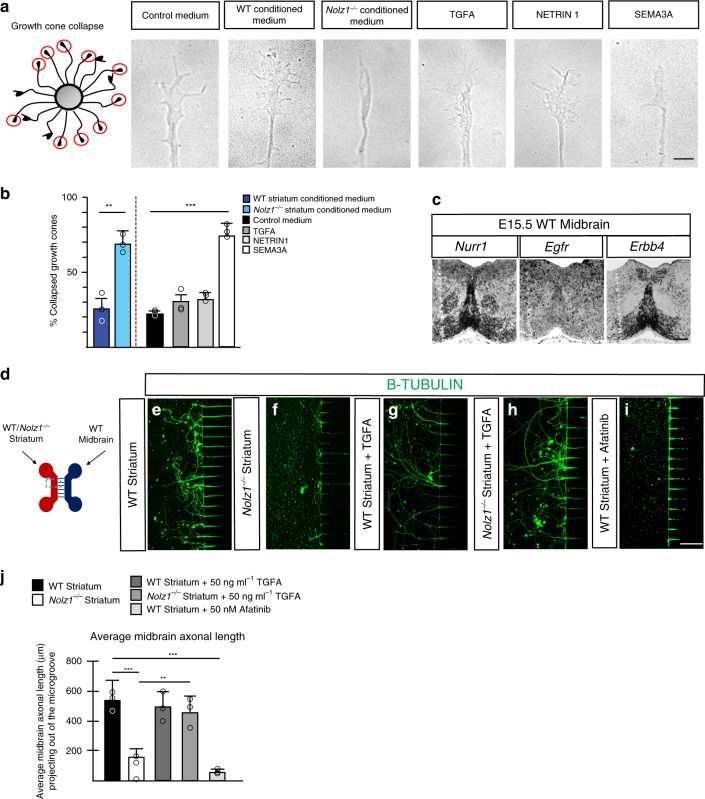
Fig. 7. Nolz1−/− mutant striatum repulses DA axons.
a Schematic representation of scoring system. Red circles indicate collapsed growth cones. Representative images of growth cones responses to control medium, E13.5 Wt and Nolz1−/− mutant-derived striatal conditioned medium, 50 ng ml−1 TGFA, 300 ng ml−1 NETRIN1 and 300 ng ml−1 SEMA3A. b Graph showing the percentage of collapsed growth cones exposed to different conditions (n = 3 independent experiments). Mean values ± standard deviation; Two-sided, unpaired T-test: Wt striatum versus Nolz1−/− striatum **p = 0.00237; Control medium versus SEMA3A medium ***p = 1.43231xE−05. c In situ hybridization showing expression of Egfr, Erbb4 and Nurr1 in E15.5 Wt ventral midbrain (coronal sections). d–i Microfluidic assay to assess attractive and repulsive effects of Wt and Nolz1−/− mutant striatal tissue and TGFA signalling on DA axons. Axons were labelled by b-TUBULIN. d Design of microfluidic platform. Primary DA neuronal cultures obtained from E13.5 Wt embryos were seeded in the cellular compartment and E13.5 Wt or Nolz1−/− mutant striatal explants were cultured in the opposing compartment. Primary DA neurons were cultured in the presence of either Wt striatal explants (e), Nolz1−/− mutant striatal explants (f), Wt striatal explants with 50 ng ml−1 TGFA (g), Nolz1−/− mutant striatal explants with 50 ng ml−1 TGFA (h) or Wt striatal explants with 50 nM Afatinib (i). j Graph showing the average axonal length of midbrain neurons projecting out of the microgroove under different conditions Mean values ± standard deviation; n = 6 biologically independent experiments; Two-sided, unpaired T-test: Wt Striatum versus Nolz1−/− Striatum ***p = 3.59072xE−06; Nolz1−/− Striatum versus Nolz1−/− Striatum + TGFA **p = 0.0017; Wt Striatum versus Wt Striatum + Afatinib ***p = 1.74752xE−06. Data are representative of three (a, c) or six (d–i) independent experiments. Scale bar (a–i): 200 μm. Source data are provided as a source data file.