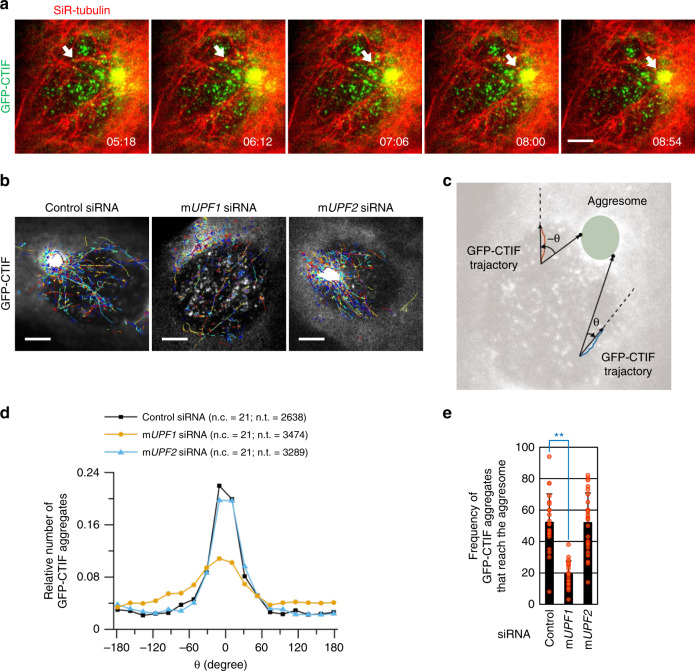
Fig. 6. UPF1 increases the frequency and fidelity of movement of CTIF aggregates.
a Live cell imaging of GFP-CTIF (green) with SiR-conjugated tubulin (red). MEFs were transiently transfected with GFP-CTIF and were treated with SiR-tubulin for staining of microtubules. Then, the cells were visualized by line-scan confocal microscopy. Scale bar, 5 μm; n = 3. b GFP-CTIF aggregates moving toward the aggresome after downregulation of mouse (m) UPF1 or mUPF2. MEFs were transfected with mUPF1 siRNA, mUPF2 siRNA, or nonspecific control siRNA. One day later, the cells were transiently transfected with GFP-CTIF. Two days later, the cells were treated with MG132 and visualized by line-scan confocal microscopy. The trajectories of individual GFP-CTIF aggregates that moved over 590 nm are depicted. Scale bar, 5 μm; n = 3. c The degree of the migration direction of individual GFP-CTIF aggregates with respect to the aggresome. We first determined the brightest position corresponding to the aggresome periphery closest to the GFP-CTIF aggregates of interest. Then, we measured the angle between the linear trajectory of the GFP-CTIF and a line connecting the brightest position with the initial point of the trajectory of GFP-CTIF. d The distribution of the observed angle values of individual GFP-CTIF aggregates. The number of the cells at each angle was normalized to the total number of the cells analyzed. The number of cells (n.c.) were 21 from three independent experiments for all cases and the number of trajectories (n.t.) were 2638 (Control siRNA), 3474 (mUPF1 siRNA), and 3289 (mUPF2 siRNA), respectively. e The frequency of GFP-CTIF aggregates that reach the aggresome. The number of GFP-CTIF aggregates (per cell) that reached the aggresome during 15 s were counted. Data are presented as mean values ± SD. Two-tailed, unequal-sample variance Student’s t test was carried out to calculate the P values. **P < 0.000. The exact P values are provided in a Source Data File; n = 23 (Control siRNA), 22 (mUPF1 siRNA), and 25 (mUPF2 siRNA) cells from three independent experiments were examined; Source data are provided as a Source Data File.