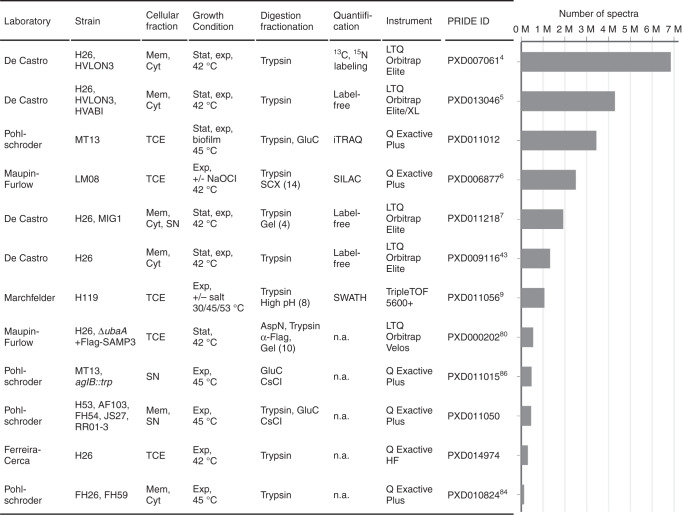
Fig. 1. Summary of ArcPP datasets comprising a total of more than 23 million spectra.
A diverse array of MS datasets for H. volcanii has been compiled for the initial reanalysis by the ArcPP. For each dataset, strains (separated by comma), cellular fractions (Mem, membrane; Cyt, cytosol; SN, culture supernatant, TCE, total cell extract), growth conditions (stat, stationary; exp, exponential growth phase), enzyme(s) used for protein digestion, and fractionation methods on peptide (SCX, strong cation exchange chromatography; high pH, high pH reversed-phase chromatography) or protein level (gel, SDS-PAGE; CsCl, CsCl gradient) with the number of fractions indicated in parentheses, quantification methods (iTRAQ, isobaric tags for relative and absolute quantitation; SILAC, stable isotope labeling with amino acids in cell culture; SWATH, sequential window acquisition of all theoretical fragment ion spectra), instruments employed, corresponding PRIDE IDs with references and the sum of all spectra are noted. Experiments were performed by five different laboratories. For more details see Supplementary Tables 2-4 and Supplementary Data 1-2. Source data are provided as a Source data file.