Figure 1.
Summary of Nanopore Sequencing Data from HM-276D and HM-277D Microbial Communities
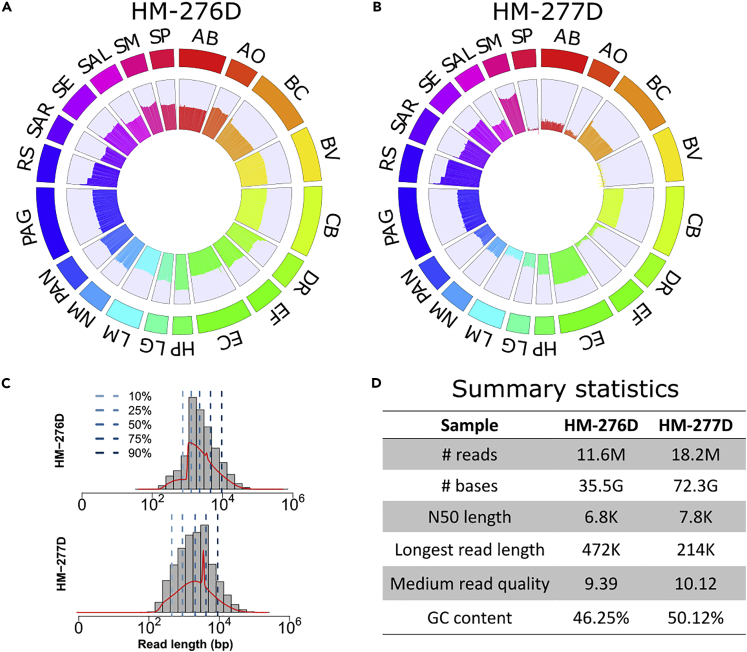
(A–C) (A and B) Circos plots of read coverage across whole genome of 20 bacterial strains from (A) HM-276D and (B) HM-277D. Each chromosome was divided into bins with 5,000 bp width. Average read coverage was calculated within each bin and converted to log scale to facilitate viewing and comparing between bacterial strains. AB, Acinetobacter baumannii; AO, Actinomyces odontolyticus; BC, Bacillus cereus; BV, Bacteroides vulgatus; CB, Clostridium beijerinckii; DR, Deinococcus radiodurans; DF, Enterococcus faecalis; EC, Escherichia coli; HP, Helicobacter pylori; LG, Lactobacillus gasseri; LM, Listeria monocytogenes; NM, Neisseria meningitides; PAN, Propionibacterium acnes; PAG, Pseudomonas aeruginosa; RS, Rhodobacter sphaeroides; SAR, Staphylococcus aureus; SE, Staphylococcus epidermidis; SAL, Streptococcus agalactiae; SM, Streptococcus mutans; SP, Streptococcus pneumonia. (C) Read length distribution of HM-276D and HM-277D datasets. Blue dashed lines represent different quantiles. Red line represents the density of read length distribution.
(D) Summary statistics of HM-276D and HM-277D datasets. Each value was calculated by using pycoQC (Leger and Leonardi, 2019) and LongreadQC. Real-time statistics are shown in Figures S1–S5.