Figure 5.
PRMT1 Is Required for Progesterone-Dependent Expression of a Subset of PR Target Genes
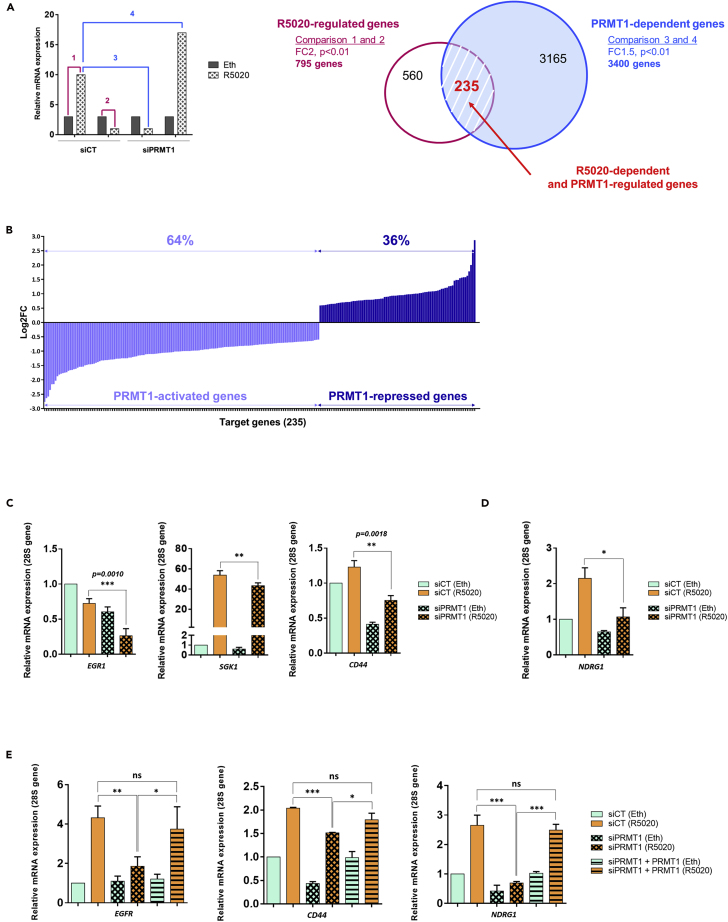
(A) Genome-wide RNA-seq analysis was performed using T47D cells to identify genes dependent on PRMT1 for R5020-regulated expression. Left panel: Hypothetical results of gene expression profiles for a given gene, illustrating how specific pairwise comparisons were performed between datasets for individual samples. Numbered bars represent hypothetical mRNA levels from RNA-seq data for cells expressing the indicated siRNAs (PRMT1 or CT) and treated for 6 h with ethanol (Eth) or R5020 (10 nM). Colored numbers represent pairwise comparisons performed to determine sets of genes for which mRNA levels were significantly different between the samples. For instance, comparison 1 = set of R5020-regulated genes (fold change ≥2, adjusted p < 0.01); comparison 4 = set of PRMT1-dependent genes (fold change >1.5, adjusted p < 0.01). Right panel: Pink and white Venn diagram represents the R5020-regulated genes in cells expressing siCT (comparisons 1 and 2); blue Venn diagram for PRMT1-dependent genes in R5020-treated cells (comparisons 3 and 4). Overlap area (in red) indicates the number of genes shared among sets. Controls for T47D cell treatments are provided in the Figures S4A–S4C.
(B) Representation of fold changes (log2FC) of all target gene expressions identified by RNA-seq analysis (235 genes). On the left (light blue), genes that are downregulated with siPRMT1 (64%), thus positively regulated by PRMT1. On the right (dark blue), genes that are negatively regulated by PRMT1 (36%).
(C and D) T47D cells were transfected with siRNAs against PRMT1 (or control) and treated for 6 h with ethanol (Eth) or R5020 (10 nM). Total RNA was prepared and cDNAs were analyzed by RT-qPCR with the indicated primers.
(E) cDNAs of T47D cells transfected with anti-PRMT1 siRNAs and co-transfected with a plasmid expressing a rat-PRMT1, then treated for 6 h with R5020 (10 nM) or vehicle ethanol (Eth), were analyzed by RT-qPCR.
All the cDNA mean values were normalized against the expression of 28S ribosomal mRNA as reference. Results shown are mean ± SEM for, at least, three independent experiments. The p value was calculated using a paired t test: ∗ indicates p ≤ 0.05, ∗∗p ≤ 0.01 and ∗∗∗p ≤ 0.001.