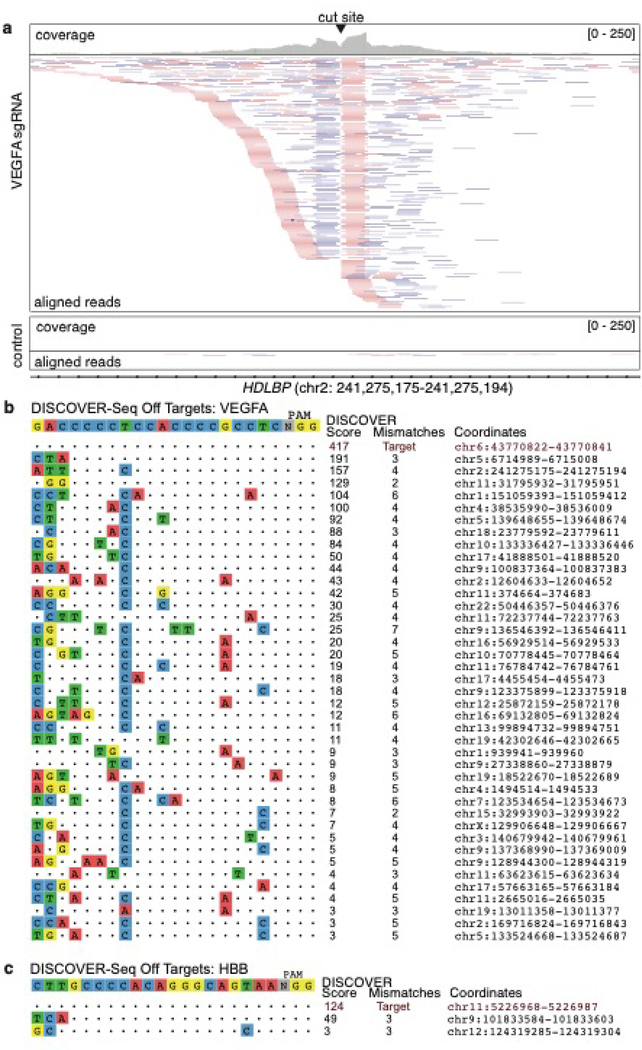
Figure 4: Example output of a successful DISCOVER-Seq experiment.
(a) Snapshot of aligned bam files in IGV from K562 cells edited with VEGFA site 2 gRNA (top) and non-targeting gRNA (bottom, control). Shown is one of the off-target sites for the VEGFA site 2 gRNA. The top track for each sample shows the read coverage while the bottom track shows the aligned reads. Forward reads are shown in pink, reverse reads are shown in purple and grey lines connect read pairs. Many reads start and end in the cut site (centered). The control sample displays the level of noise that can be expected from MRE11 pulldown in unedited cells. (b) Example BLENDER output showing the DISCOVER-Seq hits in K562 cells edited with VEGFA site 2 gRNA and Cas9 RNP. The intended target sequence is shown on top and identified sites are ordered from top to bottom by DISCOVER score. Identity to the intended target is shown as dots, mismatches are indicated by colored nucleotides, and the hg38 genomic coordinates of each off-target are listed on the right. (c) Example BLENDER output showing the DISCOVER-Seq hits in K562 cells edited with HBB gRNA and Cas9 RNP.