Fig. 1.
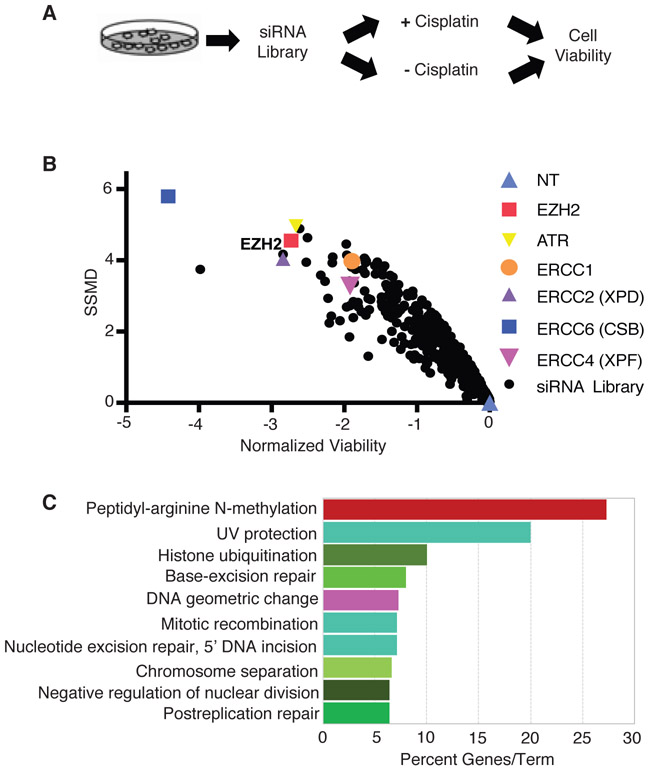
A siRNA screen targeting nuclear enzymes identifies genes that mediate cisplatin resistance in SCLC. a Primary screen format: H128 cells were transfected with a siRNA library biased towards targeting nuclear enzymes. 48 hours post-transfection, cells were treated with or without 10 μM cisplatin for 72 hours prior to measuring cell viability. b Primary screen results: the normalized cell viability was plotted against the strictly standardized mean difference (SSMD) for each gene targeted by the library. Normalized viability was calculated as the log2 ratio of treated versus untreated cell viability relative to the non-targeting (NT) siRNA control. c Summary of most enriched categories from Gene Ontology (GO) analysis of cisplatin sensitization hits (n = 118). The top 10 significant processes (p ≤ 0.01) are shown, sorted by the percent associated genes found.