Figure 2. ERK signaling promotes the integration of newly synthesized purines into nucleic acids.
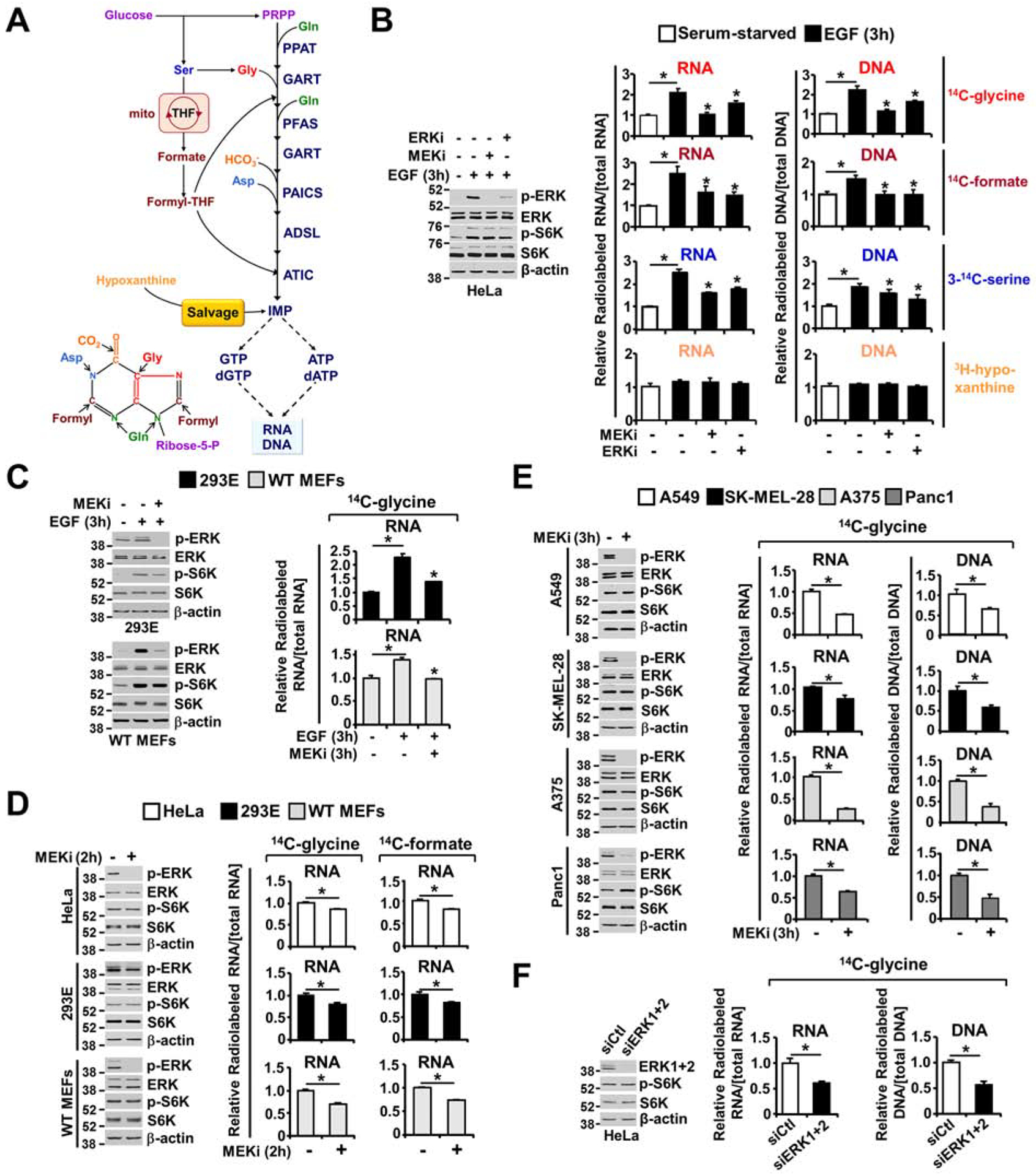
(A) Schematic of the purine synthesis pathways, including the pentose phosphate pathway, serine biosynthesis pathway and the mitochondrial THF cycles providing carbon and nitrogen for de novo purine nucleotide synthesis. Premade nucleobases, such as hypoxanthine, can sustain nucleotide synthesis via the purine salvage pathway. (B) Immunoblots assessing ERK and mTORC1 signaling; the relative incorporation of 14C from glycine, formate, and serine; and the relative incorporation of 3H from hypoxanthine into RNA and DNA. Labeling was performed for 3 hours under serum-free or EGF (50 ng/ml, 3 hours) stimulation conditions in the presence or absence of U0126 (MEKi, 10 μM) or SCH772984 (ERKi, 1 μM), reflecting de novo purine synthesis (14C-glycine, 14C-formate), one-carbon metabolism into purine nucleotides (3-14C-serine) and purine salvage pathway activity (3H-hypoxanthine) (see also Figure S2B and S2C). (C) HEK293E cells and MEFs were treated as in (B) but labeled with only 14C-glycine for 3 hours. The relative levels of incorporation of 14C from glycine into RNA are shown. Immunoblots performed in parallel to the radio-tracing experiments are shown. (D) As in (B, C), but the cells were cultured in 10% dialyzed serum and treated with vehicle or U0126 (MEKi, 10 μM, 2 hours) and labeled with 14C-glycine for 2 hours. (E) Relative incorporation of 14C-glycine into RNA, with labeling for 3 hours under serum-free conditions in the given cancer cell lines with high levels of ERK signaling (A549, SK-MEL-28, A375, and Panc1), treated with vehicle or U0126 (MEKi, 10 μM) for 3 hours (see also Figure S2D and S2E). (F) As in (D), HeLa cells were transfected with ERK1 and ERK2 siRNAs or nontargeting controls (siCtl) for 48 hours. Cells were cultured in 10% dialyzed serum for 15 hours and were then treated with vehicle or U0126 (MEKi, 10 μM, 2 hours) and concurrently labeled with 14C-glycine for 2 hours. The data are graphed as the means ± SDs of biological triplicates and are representative of at least two independent experiments (B-F). * P<0.05 by one-way ANOVA with Tukey’s post hoc test for multiple pairwise comparisons (B, C) and two-tailed Student’s t test for pairwise comparisons (D, E, F).
