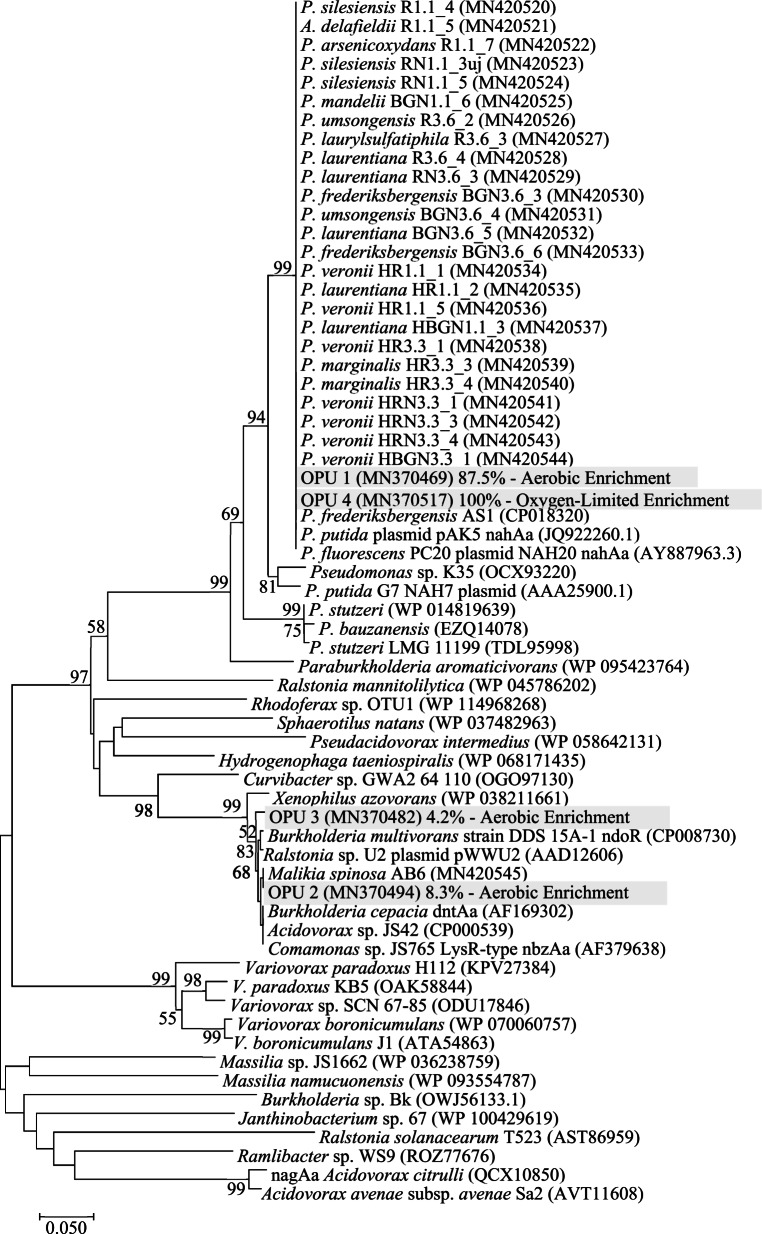
Fig. 4.
Phylogenetic analysis of NDO-related 2Fe-2S reductase protein sequences using the Neighbor-Joining method (Saitou and Nei 1987). The percentage of replicate trees in which the associated OPUs clustered together in the bootstrap test (1000 replicates) is shown next to the branches (Felsenstein 1985). The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Poisson correction method (Zuckerkandl and Pauling 1965) and are in the units of the number of amino acid substitutions per site. The analysis involved 66 amino acid sequences. All positions containing gaps and missing data were eliminated. There were a total of 222 positions in the final dataset. Evolutionary analyses were conducted in MEGA7 (Kumar et al. 2016)