Figure 6.
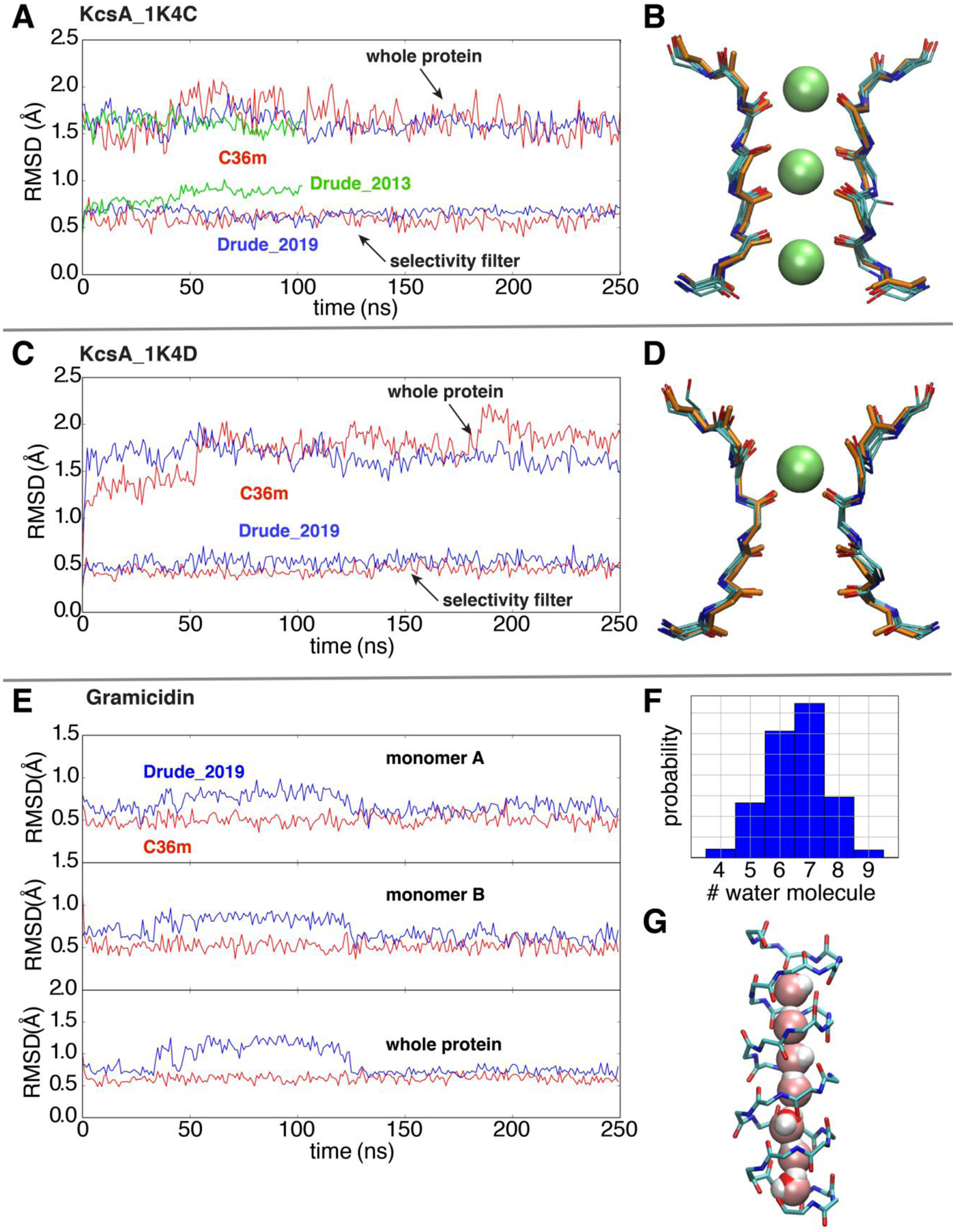
Simulations of ion channels to validate the optimized Drude 2019 FF. (A) and (C) RMSD analyses based on the backbone non-hydrogen atoms of the whole KcsA channel and the selectivity filter in simulations, respectively, using the additive C36m (red), Drude-2019 (blue) and Drude-2013 (green) FFs. The references for backbone RMSD measurements are the initial crystal structures 1K4C (conductive) and 1K4D (constricted). (B) and (D) The overlay of 5 snapshots (one frame for every 50ns in the 250ns trajectory) and the crystal structures (orange) of the selectivity filter. (E) Time series of the backbone RMSD of two monomers (top and middle) and the whole protein (bottom) of gramicidin (reference PDB: 1JNO120). (F) The histogram of the number of water molecules in gramicidin during the whole trajectory. (G) A representative snapshot with a single-file water wire in the gramicidin A channel.
