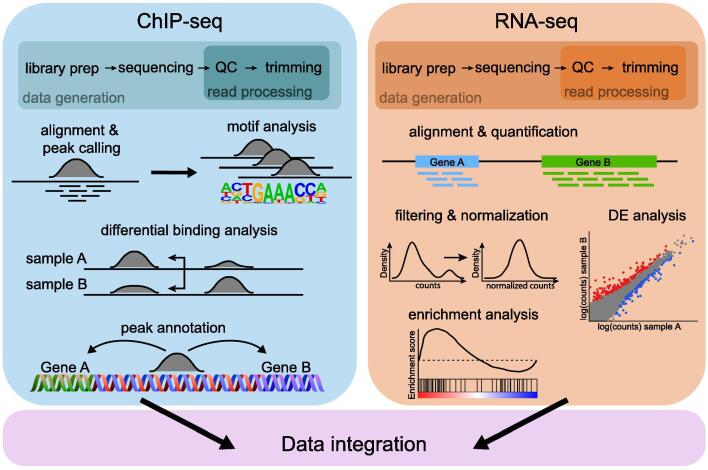
Fig. 3.
Standard processing workflow of ChIP-seq and RNA-seq. In both cases, the quality of the sequenced reads is checked before performing the alignment. The ChIP-seq data analysis continues with peak calling, followed by differential binding analysis. Searching for motifs in the peak regions and peak annotation are crucial steps. For RNA-seq, the aligned reads are quantified at gene level, the raw counts are then filtered and normalized to enable further comparisons. The differential expression analysis provides a list of significant genes, from which biological meaning may be retrieved. QC: Quality control, DE: differential expression.