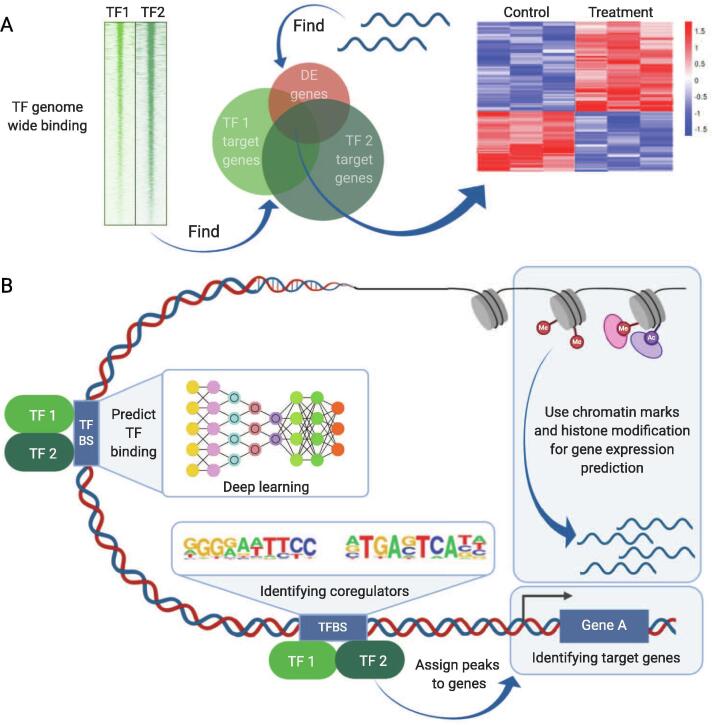
Fig. 4.
Data integration approaches. (A) ChIP-seq and RNA-seq data can be integrated in a discretized fashion by determining the overlap of significantly affected genes in the 2 assays. (B) Newer approaches combine ChIP-seq data from multiple TFs and HMs together with expression data and accessibility data such as DNase-seq and ATAC-seq. They achieve data integration through various different mathematical concepts such GLMs, HMMs and deep neural networks to identify co-regulators, predict gene expression or model TF binding. DE: differential expression, TF: transcription factor. This figure was created with BioRender (biorender.com).