Fig. 1.
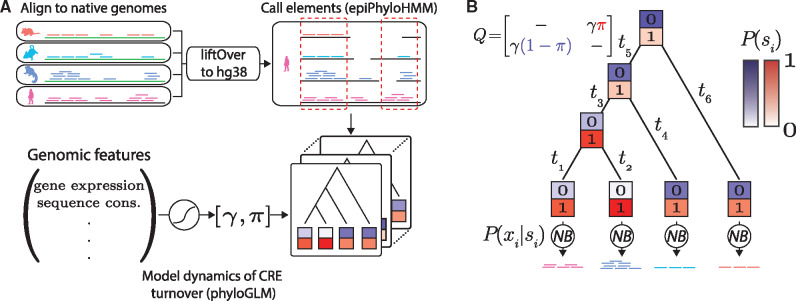
Illustration of modeling framework. (A) ChIP-seq data are aligned separately to the reference genome for each species then converted to the coordinate system of the human (hg38) genome using the liftOver program (see Materials and Methods). Only regions of apparent one-to-one orthology are considered, based on synteny. Cis-regulatory elements (CREs) that are active in one or more species are then identified using epiPhyloHMM. Finally, the dynamics of CRE turnover within these elements are modeled using phyloGLM, which accounts for the associations between various genomic features and local rates of gain and loss. (B) Both epiPhyloHMM and phyloGLM use a core “two-state” phylogenetic model in which the presence (si = 1) and absence (si = 0) of CREs are allowed to change in a branch length-dependent manner along a fixed phylogeny, according to a continuous-time Markov model. The model is defined by an instantaneous rate matrix Q (dashes indicate values required for rows to sum to zero). The conditional probabilities of the raw ChIP-seq read counts at the tips of the tree (xi) given the corresponding state (si) are modeled using negative binomial (NB) distributions. The color intensities for the “0” and “1” boxes are proportional to the probability of each state. π, stationary frequency of CRE presence; γ, gain/loss rate; ti, length of branch i.