Fig. 3.
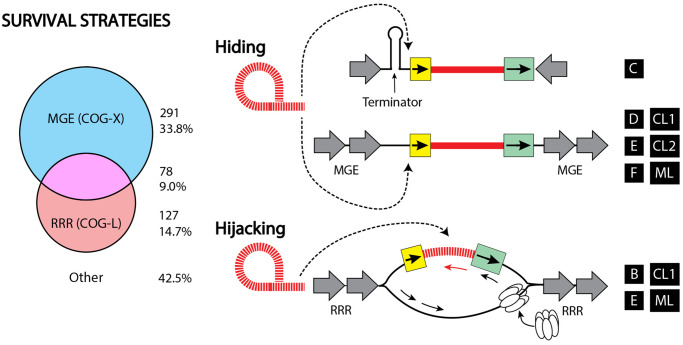
Group II introns exhibit both hiding and hijacking survival strategies. Left shows a Venn diagram describing the number of gII introns containing at least one neighbor belonging to MGE (COG-X) or RRR (COG-L) categories, which are suggestive of hiding and hijacking strategies, respectively. Right shows a conceptual depiction of these strategies. Class C, the most abundant class of gII introns in bacteria, hide by inserting downstream of transcription terminators. Other classes of gII introns (CL1, CL2, D, E, F, ML) hide in genomes by avoiding key functions and colocalizing with other mobile genetic elements (MGEs). An alternative strategy that some classes utilize (B, CL1, E, ML) is to hijack the functions of their genomic neighborhoods, timing their expression with conditions that are conducive to gII intron proliferation, such as ssDNA and replication, recombination, and repair functions (RRR). Intron is in red (dashed lines RNA, solid lines DNA). Gray arrows are flanking features as in figure 2.