Significance
All plants and animals are associated with communities of viruses and microbes that interact via a suite of metabolites. These components play critical roles in the success of these assemblages; however, the role of individual components (i.e., bacteria, viruses, metabolites) and how these govern ecological interactions between macroorganisms is not understood. This study investigates the role of holobiont components in coral–turf algal interactions. The data demonstrate that an emergent microbiome and metabolome form at the interface between coral and turf algae in competitive interactions. Machine learning analyses show that this emergent community predicts the outcome of these interactions. These results provide insight into rules of community assembly in microbiomes and the roles of holobiont components in mediating ecological interactions.
Keywords: holobiont, metabolomics, metagenomics, microbial ecology, coral reefs
Abstract
Viruses, microbes, and host macroorganisms form ecological units called holobionts. Here, a combination of metagenomic sequencing, metabolomic profiling, and epifluorescence microscopy was used to investigate how the different components of the holobiont including bacteria, viruses, and their associated metabolites mediate ecological interactions between corals and turf algae. The data demonstrate that there was a microbial assemblage unique to the coral-turf algae interface displaying higher microbial abundances and larger microbial cells. This was consistent with previous studies showing that turf algae exudates feed interface and coral-associated microbial communities, often at the detriment of the coral. Further supporting this hypothesis, when the metabolites were assigned a nominal oxidation state of carbon (NOSC), we found that the turf algal metabolites were significantly more reduced (i.e., have higher potential energy) compared to the corals and interfaces. The algae feeding hypothesis was further supported when the ecological outcomes of interactions (e.g., whether coral was winning or losing) were considered. For example, coral holobionts losing the competition with turf algae had higher Bacteroidetes-to-Firmicutes ratios and an elevated abundance of genes involved in bacterial growth and division. These changes were similar to trends observed in the obese human gut microbiome, where overfeeding of the microbiome creates a dysbiosis detrimental to the long-term health of the metazoan host. Together these results show that there are specific biogeochemical changes at coral–turf algal interfaces that predict the competitive outcomes between holobionts and are consistent with algal exudates feeding coral-associated microbes.
Coral and algae holobionts are assemblages of the macroorganisms and their associated viruses, bacteria, archaea, and protists (1). The microbial portion of the holobiont, the microbiome, is often species-specific, temporally stable, and distinct between microhabitats within the host (2–9). The microbiome performs a wide array of functions that influence both host physiology (10–12) and the biogeochemical cycling of matter and energy (13–22). The health of the holobiont is linked to the composition of its microbial constituents, which can be disrupted by various stressors and lead to dysbioses (23, 24). Host-associated microbes may also increase holobiont resistance and resilience to both local and global stressors (25). For instance, viral and bacterial symbionts may ward off potential pathogens through lysis, niche exclusion, and production of antibiotics (26–28). These interacting biological entities will influence the chemistry of holobionts, which will be reflected in the metabolites.
Understanding the roles of holobionts in ecosystem function has become increasingly important as many reefs that were formerly dominated by coral have been shifting to systems dominated by turf and fleshy macroalgae (29–33). Turf algae are among the most abundant algal competitors that corals face (34–36) and, as such, play an important role in initiating algal phase shifts on coral reefs. Competition with turf algae is known to alter the microbial communities associated with corals (37). However, the role of the microbiome and its associated metabolome in determining the outcome of these events (i.e., whether a coral wins or loses against its algal competitor) is still relatively unknown.
To examine how microbial diversity, metabolic capacity, and biochemistry affect ecological interactions between macroorganisms, both surface-associated water (n =18) and tissue (n = 36) samples were taken at a centimeter-scale spatial resolution across in situ coral–turf algal interactions from reefs on the island of Curaçao. All of these interactions involved turf algal holobionts interacting with either Diploria strigosa or Orbicella faveolata (formerly Montastraea faveolata) corals. Tissue samples were processed for metagenomic sequencing and metabolomic profiling, and the surface-associated samples were analyzed by epifluorescence microscopy (SI Appendix, Supplementary Methods). Microbial and viral abundance in the holobiont were directly quantified through microscopy. The microbiome was taxonomically and functionally profiled through metagenomic sequencing, and the molecular composition of the holobiont was assessed by untargeted metabolomic analysis. A machine learning approach was applied to identify which bacterial taxa, functional genes, and metabolites were most important for determining whether a coral was winning (i.e., had no algal overgrowth and no visible signs of paling or necrosis) or losing (i.e., had algal overgrowth and had visible signs of paling or necrosis) against its turf algal competitor.
The results of this study show that there are specific functional genes, microbial taxa, and metabolites which distinguish coral, turf algae, and interface communities and that these functions, taxa, and metabolites are also linked to the competitive outcomes of these interactions. Specifically, the data demonstrate that there is an emergent microbial community that forms at the interface between coral and turf algae, which is characterized by larger and more numerous bacterial cells, a greater proportion of Bacteroidetes, and a lower proportion of Firmicutes, as well as an enrichment in genes involved in bacterial cell growth and division. Furthermore, the same taxa and functional genes (i.e., Bacteroidetes, Firmicutes, and bacterial cell growth and division genes) are also significant predictors of whether the corals in these interactions were winning or losing against their turf algal competitor (i.e., whether the coral in the interaction was being overgrown by algae and showed visible signs of paling and/or tissue necrosis). In sum, the data presented here provide a comprehensive multiomic analysis of in situ coral–turf algal interactions, which illustrates the ecological role of the holobiont in organismal interactions in one of the most diverse and economically valuable ecosystems on Earth: coral reefs.
Results
Site Selection and Water Chemistry.
All samples were collected from the Caribbean island of Curaçao in November 2015 (SI Appendix, Fig. S1, Table S1, and Supplementary Methods). Site-level water chemistry data including inorganic nitrogen, phosphate, and dissolved organic carbon (DOC) concentrations are shown in SI Appendix, Table S2. No sites were identified as outliers in the dataset for any of the measured water chemistry parameters, and these parameters were not significantly different between coral species (n = 6, all ANOVA P values ≥0.34) or competition outcome (n = 6, all ANOVA P values ≥0.27) (SI Appendix, Table S2).
Concentrations of Surface-Associated Viruses and Microbes Across Coral–Algal Interactions.
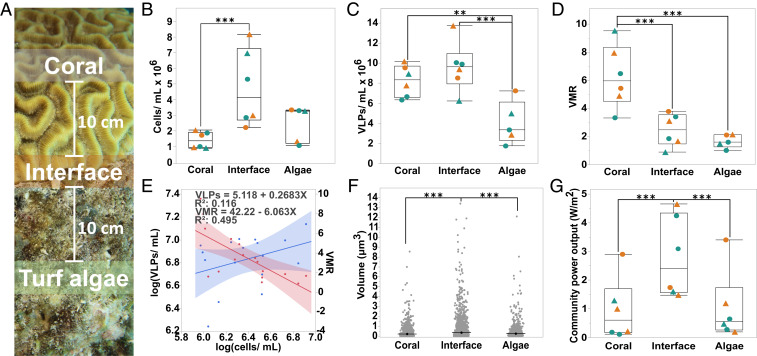
Samples for microscopic direct counts were taken by suctioning water and mucus directly off the surface of the coral, the turf algae, and the coral–turf algal interface with a syringe (Fig. 1A). There was a general trend for the interfaces (4.80 × 106 ± 2.46 × 106 cells∙mL−1; mean ± SEM) to exhibit a higher microbial abundances than corals (1.45 × 106 ± 2.12 × 106 cells∙mL−1; mean ± SEM) or turf algae (2.50 × 106 ± 5.19 × 106 cells∙mL−1; mean ± SEM (Fig. 1B). Coral-associated (8.24 × 106 ± 1.56 × 106 VLPs∙mL−1; mean ± SEM) and interface-associated (9.64 × 106 ± 2.44 × 106 virus-like particles [VLPs]∙mL−1; mean ± SEM) samples had a significantly higher concentration of viruses than turf algal-associated (4.04 × 106 ± 2.14 × 106 VLPs∙mL−1; mean ± SEM) samples (Fig. 1C). Furthermore, coral-associated (6.270 ± 2.222; mean ± SEM) samples had a significantly higher virus-to-microbe ratio (VMR) than the interface-associated (2.465 ± 1.156; mean ± SEM) or turf algae-associated (1.674 ± 0.470; mean ± SEM) samples (Fig. 1D). Overall, VLPs and microbial cells exhibited a significantly positive correlation (R2 = 0.116, P = 0.0008) with a slope of less than one (m = 0.268), and the VMR showed a significantly negative correlation (R2 = 0.495, P = 0.0016) with microbial cells (Fig. 1E), indicating a reduced lytic production of viruses at higher microbial concentrations.
Fig. 1.
Analysis of coral-, interface-, and turf algal-associated virus and microbe communities by epifluorescence microscopy. (A) Sampling schematic. Separate surface-associated water samples for microbial and viral counts and tissue biopsies for multiomics were taken from the coral, the turf algae, and the interaction interface between the two macroorganisms. Interface samples were taken at the coral–turf algae interaction zone, and coral and turf algae water and tissue samples were taken 10 cm away from the interface. All samples were taken from coral–turf algal interactions at a 10–15 m depth. (B) Concentration of microbial cells per milliliter. (C) Concentration of VLPs per milliliter. (D) VMR. (E) Linear regression analysis of the concentration of VLPs (blue line) and VMR (red line) as a function of microbial cell concentration. (F) Microbial cell size. (G) The community power output in W/m2 as predicted by MTE. For B–D and G, triangles (▲) represent coral losing interactions, circles (●) represent coral winning interactions, orange symbols represent interactions with D. strigosa, and green symbols represent interactions with O. faveolata (n = 18, **P ≤ 0.05, ***P ≤ 0.01)
Epifluorescence micrographs were also used to determine microbial cell size (SI Appendix, Supplementary Methods), and the results showed that there were significantly larger microbes (P < 0.001) associated with the coral–algal interface (0.351 ± 0.0094 μm3; mean ± SEM) when compared to the microbes associated with either the coral (0.212 ± 0.0134 μm3; mean ± SEM) or the turf algae (0.248 ± 0.0146 μm3; mean ± SEM) alone (Fig. 1F). Cell sizes were used to calculate a predicted community metabolic power output via metabolic theory of ecology (MTE). MTE calculations demonstrated that the combination of cell size and concentration yielded a significantly (P < 0.001) higher predicted community power output at the coral-algal interface (280,389.167 ± 140,518.682 W∙m−2; mean ± SEM) (Fig. 1G).
Holobiont and Interface-Specific Bacterial Taxa, Functions, and Metabolites.
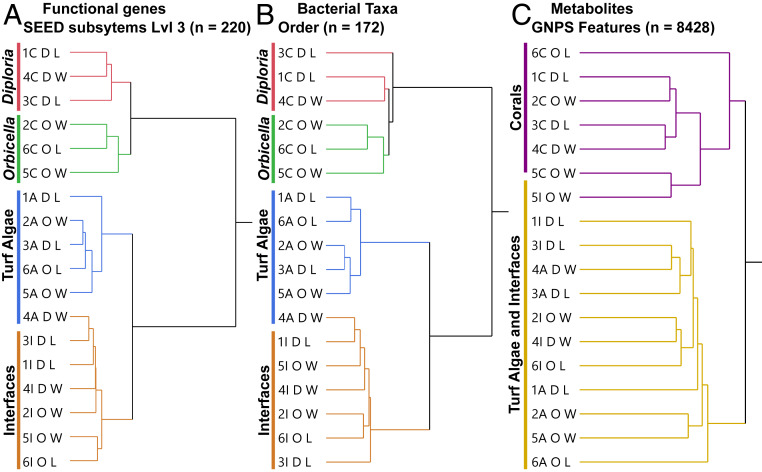
Biopsies for multiomic analysis were taken using an underwater power drill in a transect perpendicular to the coral–turf algal interface (Fig. 1A) and processed for metagenomes (see SI Appendix, Table S3 for details on metagenomic libraries) and metabolomes (SI Appendix, Supplementary Methods). Ward’s hierarchical clustering method was used to distinguish trends in the bacterial taxonomic and functional composition of metagenomes as well as for metabolomes. Overall, samples clustered by macroorganism (Fig. 2). Functional genes (Fig. 2A), bacterial taxa (Fig. 2B), and metabolites (Fig. 2C) showed the strongest support for two groups, namely, coral and noncoral (i.e., turf algae plus interface). Functional genes and bacterial taxa also showed support for two subgroups within both the coral and noncoral clusters. Within the coral cluster, the two different species of coral studied, D. strigosa and O. faveolata, were observed as distinct groups. Within the noncoral cluster, there was a subclustering of interaction interfaces versus turf algae (Fig. 2 A and B). These trends were recovered from the functional annotations at both level 1 (SI Appendix, Fig. S2) and level 3 (Fig. 2A and SI Appendix, Fig. S3) of the SEED database (38), and the taxonomic trends were observed at both the phylum (SI Appendix, Fig. S4) and order (Fig. 2B and SI Appendix, Fig. S5) levels. No trends were observed in clustering by sample site.
Fig. 2.
Hierarchical clustering of samples. Metagenomic and metabolomic data from coral, turf algae, and interface tissue samples were clustered using Ward’s hierarchical clustering method based on the functional genes (A), bacterial taxa (B), and metabolites (C). The branch tips are labeled to describe the site number (1–6), the sample type (A, algae; C, coral; I, interface), the type of coral (D = D. strigosa, O = O. faveolata), and whether the coral in the interaction was winning (W) or losing (L).
Two-way cluster analysis showed that the interaction interface group was not merely a mix of the coral and algal groups, but rather had its own unique functional, taxonomic, and metabolomic profiles (SI Appendix, Figs. S2–S6); however, the interface samples were more similar to the algal samples than to coral samples. The unique microbiome at the interface had a significantly higher relative abundance of Bacteroidetes, specifically those in the order Flavobacteriales (P < 0.0001), and a significantly lower relative abundance of Firmicutes (P = 0.003) when compared to the coral or turf algal microbiome (SI Appendix, Fig. S7). Functional gene annotations demonstrated that there was a significant overrepresentation of genes involved in cell cycle and cell division (P < 0.0001) and a significant underrepresentation of genes involved in protein metabolism (P = 0.006) at the interface (SI Appendix, Fig. S8).
In the metabolomic dataset, 182 molecules were significantly more abundant at the interface relative to both coral and turf algae. All of these molecules were unknown (i.e., there were no spectral matches to the reference libraries) except for ceramide 18:1/16:0 (P = 0.0039; Fig. 3A). This was a level 2 annotation according to the 2007 metabolomics standards initiative (39–41) (SI Appendix, Fig. S9). Ceramide 18:1/16:0 and its less saturated form, ceramide 18:2/16:0 (level 2) were found in coral and interface samples but were completely absent in turf algal samples. The less saturated (18:2/16:0) form was found in equal abundance in both coral and interface, whereas the more saturated form (18:1/16:0) was found to be significantly higher at the interface (Fig. 3 A and B and SI Appendix, Fig. S10).
Fig. 3.
Metabolites across the coral-algal interface. The ceramide 18:1/16:0 abundance (A), the less saturated form, ceramide 18:2/16:0 (B) represented as the area under the curve (AUC). The average nominal oxidation state of carbon (NOSC) (C), and the average Gibbs free energy of carbon oxidation (∆GoCox) (D). (n = 18, ***P < 0.01). Triangles (▲) represent coral losing interactions and circles (●) represent coral winning interactions. Orange symbols represent interactions with D. strigosa and green symbols represent interactions with O. faveolata.
To further investigate the metabolomic samples, each metabolite was assigned a nominal oxidation state of carbon (NOSC) and a Gibbs free energy of carbon oxidation under standard conditions (∆GoCox) based on their putative molecular formula (with mass accuracy of <1 ppm) (SI Appendix, Supplementary Methods). Overall, 7,751 of the total 8,427 features (i.e., ∼92%) were assigned a putative molecular formula using SIRIUS4, with a mass accuracy cutoff of 0.0020 ppm. Metabolites in turf algal samples had significantly lower NOSCs and significantly higher ∆GoCox values than did interface or coral samples (NOSC P = 0.0486; ∆GoCox P = 0.0201) (Fig. 3 C and D). This indicates that the biochemical compounds in turf algal samples were more reduced and were thus more energy rich. No difference in NOSC or ∆G was found between interactions where corals were winning or losing the competition with turf algae.
Machine Learning to Identify Bacterial Taxa, Functions, and Metabolites that Best Predict Competition Outcomes.
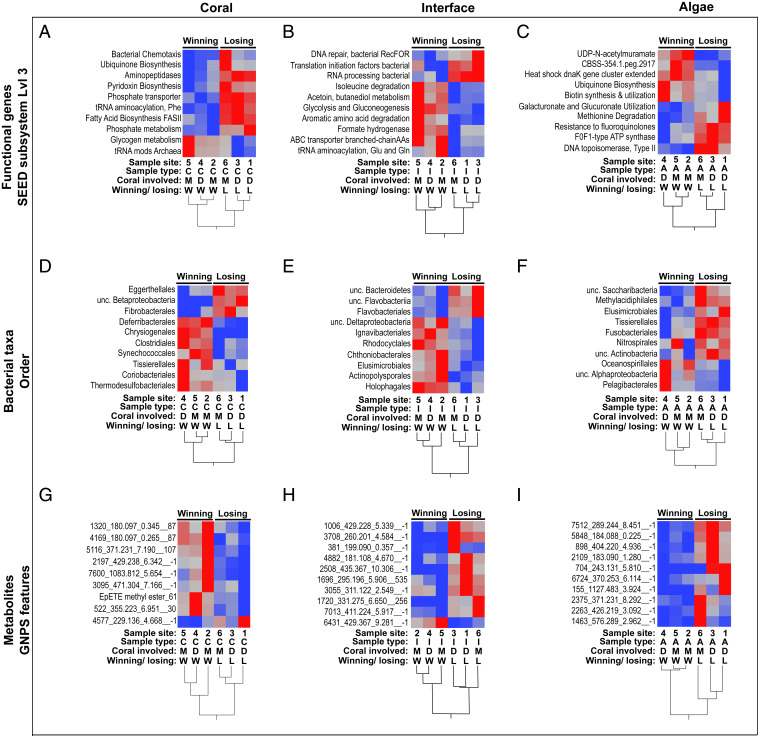
Coral–turf algal interactions were classified as winning (i.e., coral winning) or losing (i.e., coral losing) based on the criteria proposed in Barott et al. (35). Briefly, losing corals were classified as those corals that had significant algal overgrowth along with visible paling and/or tissue necrosis, whereas winning corals were corals that did not have algal overgrowth and did not show any signs of paling or necrosis. To distinguish which holobiont variables were linked to the competitive outcomes of the interactions, a random forests classification analysis was used. Random forests analysis showed that there were important functional genes (SI Appendix, Figs. S11–S13), bacterial taxa (SI Appendix, Figs. S14–S16), and metabolites (SI Appendix, Figs. S17–S19), which distinguished winning and losing interactions. The top 10 most important variables in each analysis (SI Appendix, Figs. S11–S19) were used to construct a two-way hierarchical dendrogram (Fig. 4) to visualize distinguishable groups of winning and losing corals, interfaces, and algae. Using these important taxa (Fig. 4 D–F and SI Appendix, Table S4), functional genes (Fig. 4 A–C and SI Appendix, Table S5), and metabolites (Fig. 4 G–I), samples clustered significantly by winning or losing interactions. Notably, all of the bacterial orders associated with losing interfaces were from a single bacterial phylum, Bacteroidetes.
Fig. 4.
Two-way clustering of winning and losing interactions. Samples were clustered with Ward’s hierarchical clustering method using the top random forest predictors for winning and losing corals (A, D, and G), interfaces (B, E, and H) and algae (C, F, and I) using functional genes (A–C), bacterial taxa (D–F), and metabolites (G–I) as inputs for the random forests. Abbreviations are as follows: A, algae; C, coral; D, D. strigosa; I, interface; L, losing; M, O. faveolata; W, winning. For metabolites, all of the significant predictors of the coral–turf algal competition outcome were unannotated in the GNPS database. Thus, the first number is the GNPS cluster index, the following number is the mass-to-charge ratio, the third number is the retention time, and finally, the network subcluster ID where −1 indicates a single looped compound. For all heat maps, redder indicates relatively higher abundances and bluer indicates relatively lower abundances.
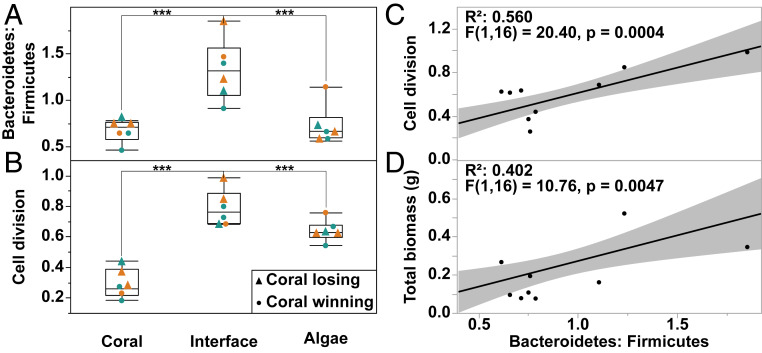
To further elucidate the taxonomic and functional mechanisms involved in winning and losing coral–algal interactions, one-way ANOVAs were performed on all bacterial phyla and level 1 SEED metabolic categories followed by Tukey post hoc analysis on all significant variables (ANOVA P values ≤0.05). This analysis revealed that there were only two phyla and two SEED subsystems that were significantly different at the interface relative to both coral and turf algal samples (i.e., ANOVA P value ≤0.05 and Tukey P value ≤0.05). The phylum Bacteroidetes was significantly enriched in interface samples, while the phylum Firmicutes was significantly depleted in the interface samples (Fig. 5A and SI Appendix, Fig. S7). Functionally, genes involved in cell division were significantly enriched in interface samples (Fig. 5B and SI Appendix, Fig. S8). The taxonomic shift toward Bacteroidetes and concomitant enrichment in cell division-related genes, as observed at the interface, was also observed on coral tissue of losing colonies (Fig. 5A and SI Appendix, Fig. S7). These results demonstrate a significantly higher Bacteroidetes-to-Firmicutes ratio at the interface and in losing corals (Fig. 5A). Regression analysis showed that the Bacteroidetes to Firmicutes ratio was significantly correlated with genes involved in cell division (Fig. 5C) (R2 = 0.560, F(1,16) = 20.4, P = 0.0004) as well as being significantly correlated with total microbial biomass (Fig. 5D) (R2 = 0.402, F(1,16) = 10.76, P = 0.0047). Taken together, these results suggest that an increase in Bacteroidetes is linked to an increase in cell division, greater bacterial cell size, and total biomass and that this shift toward faster growing, larger Bacteroidetes is linked to whether corals win or lose in their competitive interactions against benthic algae.
Fig. 5.
Results of ANOVA and subsequent regression analysis. Bacteroidetes-to-Firmicutes ratio (A), genes involved in cell division (B), regression analysis of the Bacteroidetes-to-Firmicutes ratio versus genes involved in cell division (C), and total microbial biomass in grams (D). n = 18. For A and B, triangles (▲) represent coral losing interactions and circles (●) represent coral winning interactions, and orange symbols represent interactions with D. strigosa and green symbols represent interactions with O. faveolata. (***P < 0.001).
Discussion
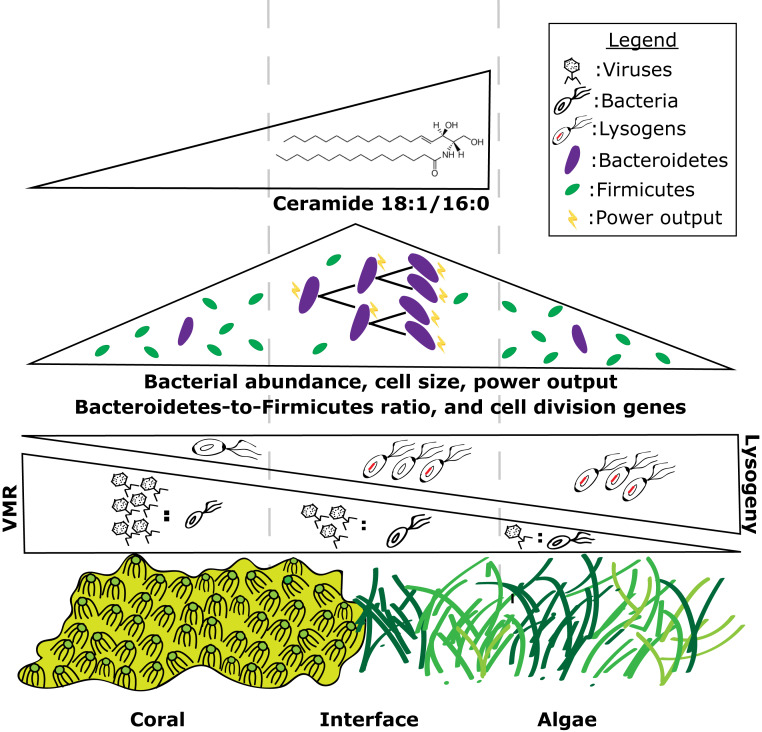
The data presented herein illustrate that there are significant differences in the size, abundance and community composition of microbes across in situ coral–turf algal interfaces (Fig. 1). These differences show there is an emergent community that forms at the interface between coral and turf algae, which is characterized by larger and more numerous bacterial cells, a higher proportion of Bacteroidetes, a lower proportion of Firmicutes, an enrichment in genes involved in bacterial cell growth and division, and an increase in the potentially proapoptotic compound ceramide 18:1/16:0.
Microbial and Viral Abundances.
The highest viral abundance was in the coral holobiont. The increased viral abundance in the coral-surface holobiont may be due to the bacteriophage adherence to mucus (BAM) dynamics described in Barr et al. (42). BAM dynamics imply that bacterial viruses (bacteriophage or phage) adhere to mucus glycoproteins through noncovalent interactions with capsid proteins. Corals may use these mucus-attached phages to defend against invading bacterial pathogens. The combination of the differences in viral and bacterial abundances leads to a significant difference in the VMR across the interface, where coral holobionts have the lowest microbial load but the highest VMR (Fig. 1D). Furthermore, metagenomic analyses demonstrated higher levels of prophage in algal samples (SI Appendix, Fig. S20), where VMR was the lowest (Fig. 1D). Taken together, these data suggest a trend toward a decrease in lytic activity and an increase in viral lysogeny at higher microbial concentrations (Fig. 1E and SI Appendix, Fig. S20). A similar trend has been seen in other environments (43, 44), including the water column of tropical coral reefs, and has been described as Piggyback-the-Winner dynamics (43). Piggyback-the-Winner posits that when VMRs are low, such as in interface and turf algal samples, there are more bacterial cells harboring lysogenic phage. This means that the bacterial assemblages at the interface and turf algae harbor a higher proportion of phage-encoded genes, which has been linked to increased pathogenicity of the overall community (45–50). These microbe-phage dynamics may be another mechanism at play in the complex interactions of coral and algal holobionts.
Microbial Biomass and Energetics.
The results also demonstrate that there are significantly larger microbial cells at the interface between coral and turf algae (Fig. 1F). This change in cell size coupled to the cell concentrations leads to a higher predicted metabolic power output at the coral–algal interface (Fig. 1G). This trend of higher power output has also been reported using calorimetry in controlled laboratory experiments (22). A higher microbial power output at the coral–algal interface means that the microbial populations here are using energy at a faster rate and are dissipating more of that energy as heat (22, 51). The increase in metabolic rate at the interface may be responsible for the reported decreased oxygen levels at the interface (22, 52–55). Understanding the direct links between bacterial taxa, cell size, power output, and biological oxygen demand may provide a more complete conceptual model of the way bacterial metabolism is involved in competitive interactions between benthic macroorganisms.
Emergent Microbiome and Metabolome at the Coral–Turf Algal Interface.
Metabolomic samples showed clear differences between coral and noncoral holobionts. The interface exhibited a unique chemical signature, however, the metabolites driving differences at the interface were mostly unknown compounds. One known compound (level 2 according to the metabolomics standards initiative) was the potentially proapoptotic molecule, ceramide 18:1/16:0 (Fig. 3A). Other bioactive lipids and proapoptotic inflammatory modulators have previously been shown to play a role in the coral holobiont (56–58), suggesting that nonself-recognition among some of the oldest extant holobionts (i.e., corals) involves bioactive lipids identical to those in highly derived taxa like humans. The data here further strengthen the hypothesis that major players of the immune response evolved during the pre-Cambrian era (59). Furthermore, turf algal metabolites were found to have more negative nominal oxidation states of carbon and higher ∆G of carbon oxidation (Fig. 3 C and D), suggesting that the biochemicals in the turf algal holobiont are more reduced and, thus, more energy rich. It may be the combination of naive coral microbes being exposed to high-energy turf algal compounds at the interface, which leads to the increase in size and power output of the bacterial cells at the interface (Fig. 1 F and G). The feeding of coral microbes on turf algal metabolites at the interface may also be in part responsible for the decrease in oxygen levels previously observed at the coral–algal interface (22, 53). Thus, we propose the “algae feeding hypothesis” where reduced, high-energy turf algae exudates feed interface and coral-associated microbial communities, often to the detriment of the coral animal.
The metagenomic and metabolomic data show that there are specific bacterial taxa, functional genes, and metabolites that distinguish coral, turf algae, and interfaces (Fig. 2), as well as winning and losing interactions (Fig. 4 and SI Appendix, Tables S4 and S5). Furthermore, these data indicate that the interface is not merely a mix of coral and turf algal holobionts, but rather has its own emergent signature (SI Appendix, Figs. S2–S6), which is more similar to the turf algal holobiont than the holobiont of coral (Fig. 2). Specifically, members of the Bacteroidetes clade are overrepresented at the interaction interface, while the phylum Firmicutes is underrepresented at the interface (Fig. 5 and SI Appendix, Fig. S7). A similar trend is observed in coral samples, where Bacteroidetes are enriched in losing corals and Firmicutes are depleted in losing coral samples (SI Appendix, Fig. S7). Thus, the data demonstrate that the Bacteroidetes-to-Firmicutes ratio is a significant predictor of whether a coral will win or lose in a competitive interaction with algae. The ratio of Bacteroidetes to Firmicutes is also a significant predictor of health status in other systems such as the human gut where this ratio has been linked to obesity (60) and in the human lung where it is linked to disease states in cystic fibrosis patients (61). The Bacteroidetes-to-Firmicutes ratio was a significant predictor of the amount of cell division genes and total microbial biomass in these holobionts (Fig. 5D). Studies in mice and humans have shown that the change in the Bacteroidetes-to-Firmicutes ratio can have significant impacts on energy output and biomass of microbial communities, with Bacteroidetes having an increased capacity to harvest energy from reduced compounds (62). Given that the abundance of Bacteroidetes and the Bacteroidetes-to-Firmicutes ratio was higher at the interface and in losing coral samples, we propose a working model (Fig. 6) whereby the reduced metabolites released by turf algae select for an increase in Bacteroidetes relative to Firmicutes, which, in turn, leads to a faster growing bacterial community with larger cells and higher energy use rate. These fast-growing microbes can outcompete the corals for resources such as oxygen, which weaken the coral and lead to eventual algal overgrowth. This link between the energy content of algal metabolites, bacterial taxonomic composition, community metabolism, and coral health provides interesting insight into the ways that different components of the holobiont affect the outcome of ecological interactions and eventually shape entire community assemblages.
Fig. 6.
Working model of how different components of coral and turf algal holobionts mediate ecological interactions on the reef. Viruses are most abundant in the coral epibiont while bacteria are most abundant at the interface. This leads to the highest VMR in the coral holobiont and the lowest VMR in the turf algal holobiont. When coupled to the increased abundance of prophage found in the turf algal metagenomes this suggests increased lysogeny in the turf algal holobiont and increased lytic activity in the coral holobiont. Bacterial cell size, microbial metabolic power output, bacterial cell division, and the Bacteroidetes-to-Firmicutes ratio is highest at the interface. The potentially proapoptotic metabolite ceramide 18:1/16:0 is found only in the coral and interface samples but is most abundant at the interface.
Despite the current progress in the field, it is worth noting that environmental multiomics still has a long way to go. Metabolomics databases are sparsely populated in regard to environmental metabolites making annotation difficult and leaving the majority of compounds unclassified. As this work and others (e.g., ref. 63) have demonstrated the need to consider all components of the holobiont, it is clear that new methods and increased sequencing efforts will be needed to provide the amount of microbial coverage necessary to properly describe the roles of the less abundant components of the holobiont such as archaea and viruses. Thus, we highlight that future work is needed to provide more robust analyses of the coral and algal holobionts and their associated metabolites.
Conclusion Statement
Overall, this study demonstrates that there are differences in both the surface-associated microbial community and the total holobionts of coral and turf algae, and that when these organisms interact, there is an emergent interface community. We hypothesize that this emergent community is driven by the coral microbiome feeding on the energy-rich exudates released by the adjacent turf algae, a phenomenon we term the algae feeding hypothesis. The data also show that specific bacterial groups such as Bacteroidetes and Firmicutes play a role in determining the competitive outcome of coral–turf algae interaction events. However, what this role is remains an open question and will require further investigation. In sum, we emphasize the role of host-associated microbial communities in ecological processes and highlight that the holobiont plays an important part in determining the outcome of coral–turf algal interactions and overall reef health.
Methods Overview
All samples were collected from in situ coral–turf algal interactions by divers on SCUBA around the Caribbean island of Curaçao in November of 2015. Surface-associated water samples and tissue biopsies were collected to assess the surface-associated epibiont and the tissue-associated microbial communities of corals and turf algae in direct competitive interactions with one another. Surface-associated water samples were processed for microscopy using both SYBR Gold and DAPI stains to quantify the abundance of bacteria and virus-like particles as well as the size of bacterial cells. Tissue biopsies were processed for metagenomics sequencing on a MiSeq platform and untargeted metabolomics via liquid chromatography tandem mass spectrometry. A machine learning approach was applied to metagenomic and metabolomic datasets to identify which bacterial taxa, functional genes, and metabolites were most important for determining whether a coral was winning or losing against its turf algal competitor. See SI Appendix, Supplementary Methods for a detailed explanation of all methods.
Data Availability.
Metagenomic sequence data from this study has been deposited into the Sequence Read Archive under the study accession code PRJNA597953.
Metabolite library spectra files are available through the GNPS at the following link: (https://gnps.ucsd.edu/ProteoSAFe/result.jsp?task=7d1e1045428548bbad575ff12445a48c&view=advanced_view).
Supplementary Material
Acknowledgments
We thank the Waitt Institute and the crew of the Plan B for their assistance in collecting samples. We thank the National Science Foundation for Grants G00009988 (to T.N.F.R.), OCE-1538567, IOS-1848671 (to L.W.K.), and OISE-1243541 (to F.R.) We thank the Undead Group—Sandi Calhoun, Kevin Green, Sean Benler, Nate Robinett, and Greg Peters—for their reading of the manuscript and useful feedback during the writing process.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: Metagenomic sequence data from this study has been deposited into the Sequence Read Archive, https://www.ncbi.nlm.nih.gov/sra (accession no. PRJNA597953).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1915455117/-/DCSupplemental.
References
- 1.Knowlton N., Rohwer F., Multispecies microbial mutualisms on coral reefs: The host as a habitat. Am. Nat. 162 (suppl. 4), S51–S62 (2003). [DOI] [PubMed] [Google Scholar]
- 2.Bourne D. G., Morrow K. M., Webster N. S., Insights into the coral microbiome: Underpinning the health and resilience of reef ecosystems. Annu. Rev. Microbiol. 70, 317–340 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Frias-Lopez J., Zerkle A. L., Bonheyo G. T., Fouke B. W., Partitioning of bacterial communities between seawater and healthy, black band diseased, and dead coral surfaces. Appl. Environ. Microbiol. 68, 2214–2228 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Littman R. A., Willis B. L., Pfeffer C., Bourne D. G., Diversities of coral-associated bacteria differ with location, but not species, for three acroporid corals on the Great Barrier Reef. FEMS Microbiol. Ecol. 68, 152–163 (2009). [DOI] [PubMed] [Google Scholar]
- 5.Mouchka M. E., Hewson I., Harvell C. D., Coral-associated bacterial assemblages: Current knowledge and the potential for climate-driven impacts. Integr. Comp. Biol. 50, 662–674 (2010). [DOI] [PubMed] [Google Scholar]
- 6.Rohwer F., Seguritan V., Azam F., Knowlton N., Diversity and distribution of coral-associated bacteria. Mar. Ecol. Prog. Ser. 243, 1–10 (2002). [Google Scholar]
- 7.Sunagawa S., Woodley C. M., Medina M., Threatened corals provide underexplored microbial habitats. PLoS One 5, e9554 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sweet M. J., Croquer A., Bythell J. C., Bacterial assemblages differ between compartments within the coral holobiont. Coral Reefs 30, 39–52 (2011). [Google Scholar]
- 9.Hester E. R., Barott K. L., Nulton J., Vermeij M. J., Rohwer F. L., Stable and sporadic symbiotic communities of coral and algal holobionts. ISME J. 10, 1157–1169 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rosenberg E., Koren O., Reshef L., Efrony R., Zilber-Rosenberg I., The role of microorganisms in coral health, disease and evolution. Nat. Rev. Microbiol. 5, 355–362 (2007). [DOI] [PubMed] [Google Scholar]
- 11.Bourne D. G. et al., Microbial disease and the coral holobiont. Trends Microbiol. 17, 554–562 (2009). [DOI] [PubMed] [Google Scholar]
- 12.Lynch S. V., Pedersen O., The human intestinal microbiome in health and disease. N. Engl. J. Med. 375, 2369–2379 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Lesser M. P., Mazel C. H., Gorbunov M. Y., Falkowski P. G., Discovery of symbiotic nitrogen-fixing cyanobacteria in corals. Science 305, 997–1000 (2004). [DOI] [PubMed] [Google Scholar]
- 14.Wegley L., Edwards R., Rodriguez-Brito B., Liu H., Rohwer F., Metagenomic analysis of the microbial community associated with the coral Porites astreoides. Environ. Microbiol. 9, 2707–2719 (2007). [DOI] [PubMed] [Google Scholar]
- 15.Beman J. M., Roberts K. J., Wegley L., Rohwer F., Francis C. A., Distribution and diversity of archaeal ammonia monooxygenase genes associated with corals. Appl. Environ. Microbiol. 73, 5642–5647 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Siboni N., Ben-Dov E., Sivan A., Kushmaro A., Global distribution and diversity of coral-associated Archaea and their possible role in the coral holobiont nitrogen cycle. Environ. Microbiol. 10, 2979–2990 (2008). [DOI] [PubMed] [Google Scholar]
- 17.Raina J.-B., Tapiolas D., Willis B. L., Bourne D. G., Coral-associated bacteria and their role in the biogeochemical cycling of sulfur. Appl. Environ. Microbiol. 75, 3492–3501 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fiore C. L., Jarett J. K., Olson N. D., Lesser M. P., Nitrogen fixation and nitrogen transformations in marine symbioses. Trends Microbiol. 18, 455–463 (2010). [DOI] [PubMed] [Google Scholar]
- 19.Kimes N. E. et al., Temperature regulation of virulence factors in the pathogen Vibrio coralliilyticus. ISME J. 6, 835–846 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haas A. F. et al., Global microbialization of coral reefs. Nat. Microbiol. 1, 16042 (2016). [DOI] [PubMed] [Google Scholar]
- 21.Ferrier-Pagès C., Godinot C., D’angelo C., Wiedenmann J., Grover R., Phosphorus metabolism of reef organisms with algal symbionts. Ecol. Monogr. 86, 262–277 (2016). [Google Scholar]
- 22.Roach T. N. F. et al., Microbial bioenergetics of coral-algal interactions. PeerJ 5, e3423 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bourne D., Iida Y., Uthicke S., Smith-Keune C., Changes in coral-associated microbial communities during a bleaching event. ISME J. 2, 350–363 (2008). [DOI] [PubMed] [Google Scholar]
- 24.Mao-Jones J., Ritchie K. B., Jones L. E., Ellner S. P., How microbial community composition regulates coral disease development. PLoS Biol. 8, e1000345 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ziegler M., Seneca F. O., Yum L. K., Palumbi S. R., Voolstra C. R., Bacterial community dynamics are linked to patterns of coral heat tolerance. Nat. Commun. 8, 14213 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ritchie K. B., Regulation of microbial populations by coral surface mucus and mucus-associated bacteria. Mar. Ecol. Prog. Ser. 322, 1–14 (2006). [Google Scholar]
- 27.Nissimov J., Rosenberg E., Munn C. B., Antimicrobial properties of resident coral mucus bacteria of Oculina patagonica. FEMS Microbiol. Lett. 292, 210–215 (2009). [DOI] [PubMed] [Google Scholar]
- 28.Rypien K. L., Ward J. R., Azam F., Antagonistic interactions among coral-associated bacteria. Environ. Microbiol. 12, 28–39 (2010). [DOI] [PubMed] [Google Scholar]
- 29.McCook L. J., Macroalgae, nutrients and phase shifts on coral reefs: Scientific issues and management consequences for the Great Barrier Reef. Coral Reefs 18, 357–367 (1999). [Google Scholar]
- 30.McCook L., Jompa J., Diaz-Pulido G., Competition between corals and algae on coral reefs: A review of evidence and mechanisms. Coral Reefs 19, 400–417 (2001). [Google Scholar]
- 31.Hughes T. P. et al., Climate change, human impacts, and the resilience of coral reefs. Science 301, 929–933 (2003). [DOI] [PubMed] [Google Scholar]
- 32.Hughes T. P. et al., Phase shifts, herbivory, and the resilience of coral reefs to climate change. Curr. Biol. 17, 360–365 (2007). [DOI] [PubMed] [Google Scholar]
- 33.Smith J. E. et al., Re-evaluating the health of coral reef communities: Baselines and evidence for human impacts across the central Pacific. Proc. Biol. Sci. 283, 20151985 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barott K. et al., Hyperspectral and physiological analyses of coral-algal interactions. PLoS One 4, e8043 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barott K. L. et al., Natural history of coral- algae competition across a gradient of human activity in the Line Islands. Mar. Ecol. Prog. Ser. 460, 1–12 (2012). [Google Scholar]
- 36.Haas A. F., Wild C., Composition analysis of organic matter released by cosmopolitan coral reef-associated green algae. Aquat. Biol. 10, 131–138 (2010). [Google Scholar]
- 37.Barott K. L., Rohwer F. L., Unseen players shape benthic competition on coral reefs. Trends Microbiol. 20, 621–628 (2012). [DOI] [PubMed] [Google Scholar]
- 38.Overbeek R. et al., The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 33, 5691–5702 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sumner L. W. et al., Proposed minimum reporting standards for chemical analysis chemical analysis working group (CAWG) metabolomics standards initiative (MSI). Metabolomics 3, 211–221 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kauhanen D. et al., Development and validation of a high-throughput LC-MS/MS assay for routine measurement of molecular ceramides. Anal. Bioanal. Chem. 408, 3475–3483 (2016). [DOI] [PubMed] [Google Scholar]
- 41.Qu F., Zhang H., Zhang M., Hu P., Sphingolipidomic profiling of Rat Serum by UPLC-Q-TOF-MS: Application to Rheumatoid Arthritis study. Molecules 23, 1324 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barr J. J. et al., Bacteriophage adhering to mucus provide a non-host-derived immunity. Proc. Natl. Acad. Sci. U.S.A. 110, 10771–10776 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Knowles B. et al., Corrigendum: Lytic to temperate switching of viral communities. Nature 539, 123 (2016). [DOI] [PubMed] [Google Scholar]
- 44.Coutinho F. H. et al., Marine viruses discovered via metagenomics shed light on viral strategies throughout the oceans. Nat. Commun. 8, 15955 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brüssow H., Canchaya C., Hardt W.-D., Phages and the evolution of bacterial pathogens: From genomic rearrangements to lysogenic conversion. Microbiol. Mol. Biol. Rev. 68, 560–602 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brüssow H., Bacteria between protists and phages: From antipredation strategies to the evolution of pathogenicity. Mol. Microbiol. 65, 583–589 (2007). [DOI] [PubMed] [Google Scholar]
- 47.Munro J., Oakey J., Bromage E., Owens L., Experimental bacteriophage-mediated virulence in strains of Vibrio harveyi. Dis. Aquat. Organ. 54, 187–194 (2003). [DOI] [PubMed] [Google Scholar]
- 48.Aziz R. K. et al., Mosaic prophages with horizontally acquired genes account for the emergence and diversification of the globally disseminated M1T1 clone of Streptococcus pyogenes. J. Bacteriol. 187, 3311–3318 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weynberg K. D., Voolstra C. R., Neave M. J., Buerger P., van Oppen M. J. H., From cholera to corals: Viruses as drivers of virulence in a major coral bacterial pathogen. Sci. Rep. 5, 17889 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Little M., Rojas M. I., Rohwer F., “Bacteriophage can drive virulence in marine pathogens𠇍 in Marine Disease Ecology, Behringer D. C., Lafferty K. D., Silliman B. R., Eds. (Oxford University Press, 2020). [Google Scholar]
- 51.Roach T. N. F., Salamon P., Nulton J., Application of finite-time and control thermodynamics to biological processes at multiple scales. J. Non-Equilib. Thermodyn. 43, 193–210 (2018). [Google Scholar]
- 52.Silveira C. B. et al., Biophysical and physiological processes causing oxygen loss from coral reefs. eLife 8, e49114 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Haas A. F. et al., Visualization of oxygen distribution patterns caused by coral and algae. PeerJ 1, e106 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gregg A. et al., Biological oxygen demand optode analysis of coral reef-associated microbial communities exposed to algal exudates. PeerJ 1, e107 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jorissen H., Skinner C., Osinga R., de Beer D., Nugues M. M., Evidence for water-mediated mechanisms in coral-algal interactions. Proc. Biol. Sci. 283, 20161137 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Quinn R. A. et al., Metabolomics of reef benthic interactions reveals a bioactive lipid involved in coral defence. Proc. Biol. Sci. 283, 20160469 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Galtier d’Auriac I. et al., Before platelets: The production of platelet-activating factor during growth and stress in a basal marine organism. Proc. Biol. Sci. 285, 20181307 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Roach T. N. F., et al. , Metabolomic signatures of coral bleaching history. 10.1101/2020.05.10.087072 (11 May 2020). [DOI] [PubMed] [Google Scholar]
- 59.Quistad S. D. et al., Evolution of TNF-induced apoptosis reveals 550 My of functional conservation. Proc. Natl. Acad. Sci. U.S.A. 111, 9567–9572 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ley R. E., Turnbaugh P. J., Klein S., Gordon J. I., Microbial ecology: Human gut microbes associated with obesity. Nature 444, 1022–1023 (2006). [DOI] [PubMed] [Google Scholar]
- 61.Garg N. et al., Three-dimensional microbiome and metabolome cartography of a diseased human lung. Cell Host Microbe 22, 705–716.e4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Turnbaugh P. J. et al., An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444, 1027–1031 (2006). [DOI] [PubMed] [Google Scholar]
- 63.Robbins S. J. et al.; ReFuGe2020 Consortium , A genomic view of the reef-building coral Porites lutea and its microbial symbionts. Nat. Microbiol. 4, 2090–2100 (2019). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Metagenomic sequence data from this study has been deposited into the Sequence Read Archive under the study accession code PRJNA597953.
Metabolite library spectra files are available through the GNPS at the following link: (https://gnps.ucsd.edu/ProteoSAFe/result.jsp?task=7d1e1045428548bbad575ff12445a48c&view=advanced_view).