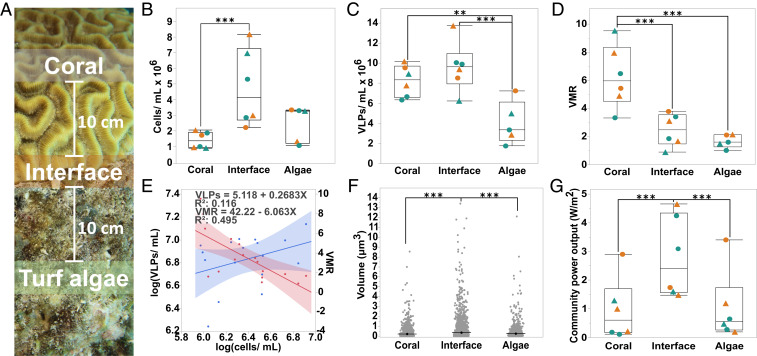
Fig. 1.
Analysis of coral-, interface-, and turf algal-associated virus and microbe communities by epifluorescence microscopy. (A) Sampling schematic. Separate surface-associated water samples for microbial and viral counts and tissue biopsies for multiomics were taken from the coral, the turf algae, and the interaction interface between the two macroorganisms. Interface samples were taken at the coral–turf algae interaction zone, and coral and turf algae water and tissue samples were taken 10 cm away from the interface. All samples were taken from coral–turf algal interactions at a 10–15 m depth. (B) Concentration of microbial cells per milliliter. (C) Concentration of VLPs per milliliter. (D) VMR. (E) Linear regression analysis of the concentration of VLPs (blue line) and VMR (red line) as a function of microbial cell concentration. (F) Microbial cell size. (G) The community power output in W/m2 as predicted by MTE. For B–D and G, triangles (▲) represent coral losing interactions, circles (●) represent coral winning interactions, orange symbols represent interactions with D. strigosa, and green symbols represent interactions with O. faveolata (n = 18, **P ≤ 0.05, ***P ≤ 0.01)