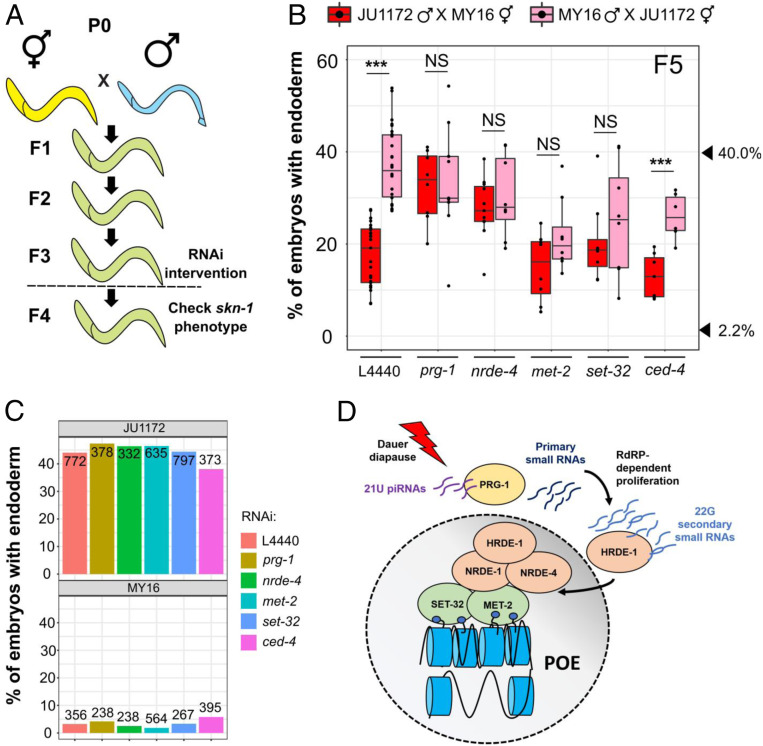
Fig. 4.
POE requires both the piRNA/nuclear RNAi pathway and factors required for H3K9me3 chromatin marks. (A) Schematic of RNAi experiments that test requirement for epigenetic regulators in POE. F3 L4 animals were treated with the indicated RNAis, and F4 L4s were used for the skn-1 RNAi assays. Animals in the control and treatment groups were siblings (Materials and Methods). (B) Knocking down prg-1, nrde-4, met-2, and set-32, but not ced-4, eliminates POE. Data points are replicates from at least two independent crosses. Boxplots represent medians, with range bars showing upper and lower quartiles. Arrowheads indicate phenotypes of JU1172 (40.0%) and MY16 (2.2%). Two-sample t test: not significant (NS), P > 0.05; ***P ≤ 0.001. (C) The effect of RNAi treatments on the skn-1(RNAi) phenotype. MY16 and JU1172 L4s were exposed to L4440 (control), prg-1, nrde-4, met-2, set-32, or ced-4 RNAi feeder strains, and the skn-1(RNAi) phenotype of the F1 were quantified. No difference between different RNAi treatments was detected (Fisher’s exact test, P > 0.05). The total number of embryos scored is indicated. (D) Model for POE (see Maintenance of POE Involves the Nuclear RNAi Pathway and Histone H3K9 Trimethylation).