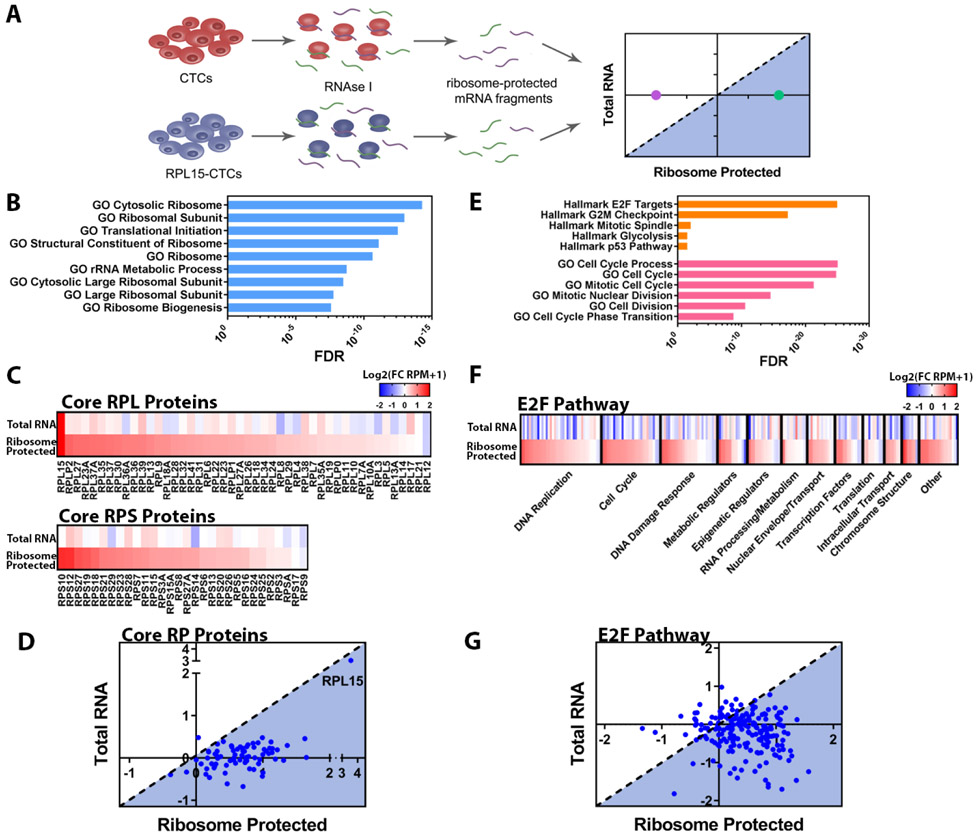
Fig. 3. RPL15 overexpression promotes translation of core ribosomal proteins and E2F pathway proteins.
(A) Schema illustrating ribosome profiling of control CTCs or RPL15-CTCs. (B) Gene Set Enrichment Analysis of transcripts preferentially bound by ribosomes in RPL15-CTCs. The most enriched ribosomal/translational GO gene sets and associated FDR values are shown. (C) Heat map representing the fold change of RPL15-CTCs relative to control for each RP gene for total RNA sequencing and ribosome profiling. Upper panel represents RPL proteins and lower panel represents RPS proteins. (D) Scatter plot representing the translational efficiency of individual RP gene transcripts. The y-axis represents the log2(fold change in RNA-seq), and the x-axis represents the log2(fold change in ribosome profiling). The shaded region represents transcripts that have increased translational efficiency relative to the level of the transcript. (E) Gene Set Enrichment Analysis of transcripts preferentially bound by ribosomes in RPL15-CTCs. The most enriched Hallmarks of Cancer gene sets from the Broad Molecular Signatures Database and most enriched cell cycle related Gene Ontology (GO) gene sets, and associated FDR values are shown. (F) Heat map representing fold change of RPL15-CTCs relative to control for each gene within the Hallmark E2F Targets gene set for total RNA sequencing and ribosome profiling. Genes were categorized according to their function and ordered based on the fold change in the ribosome profiling. (G) Scatter plot representing the translational efficiency of individual transcripts within the Hallmark E2F Targets gene set. The y-axis represents the log2(fold change in RNA-seq), and the x-axis represents the log2(fold change in ribosome profiling). The shaded region represents transcripts that have increased translational efficiency relative to the level of the transcript. Ribosome profiling was performed in duplicate.