HSV-1 ICP0 is a multifunctional immediate early protein key to effective replication in the HSV-1 lytic cycle and reactivation in the latent cycle. ICP0 transactivates gene expression by orchestrating an overall mitigation in host intrinsic/innate restrictions. How ICP0 coordinates its multiple active domains and its diverse protein-protein interactions is a key question in understanding the HSV-1 life cycle and pathogenesis. The present study focuses on delineating the regulatory effects of the SUMO-SIM interaction on ICP0 E3 ubiquitin ligase activity regarding PML II degradation. For the first time, we discovered the importance of multivalency in the PML II-ICP0 interaction network and report the involvement of different regulatory mechanisms in PML II recognition by ICP0 in HSV-1 infection.
KEYWORDS: HSV-1, ICP0, SUMO-interaction, virus-host interactions
ABSTRACT
ND10 nuclear bodies, as part of the intrinsic defenses, impose repression on incoming DNA. Infected cell protein 0 (ICP0), an E3 ubiquitin ligase of herpes simplex virus 1 (HSV-1), can derepress viral genes by degrading ND10 organizers to disrupt ND10. These events are part of the initial tug of war between HSV-1 and host, which determines the ultimate outcome of infection. Previously, we reported that ICP0 differentially recognizes promyelocytic leukemia (PML) isoforms. ICP0 depends on a SUMO-interaction motif located at residues 362 to 364 (SIM362-364) to trigger the degradation of PML isoforms II, IV, and VI, while using a bipartite sequence flanking the RING domain to degrade PML I. In this study, we investigated how the SUMO-SIM interaction regulates the degradation of PML II and PML II-associated proteins in ND10. We found that (i) the same regulatory mechanism for PML II degradation was detected in cells permissive or nonpermissive to the ICP0-null virus; (ii) the loss of a single SIM362-364 motif was restored by the presence of four consecutive SIMs from RNF4, but was not rescued by only two of the RNF4 SIMs; (iii) the loss of three C-terminal SIMs of ICP0 was fully restored by four RNF4 SIMs and also partially rescued by two RNF4 SIMs; and (iv) a PML II mutant lacking both lysine SUMOylation and SIM was not recognized by ICP0 for degradation, but was localized to ND10 and mitigated the degradation of other ND10 components, leading to delayed viral production. Taken together, SUMO regulates ICP0 substrate recognition via multiple fine-tuned mechanisms in HSV-1 infection.
IMPORTANCE HSV-1 ICP0 is a multifunctional immediate early protein key to effective replication in the HSV-1 lytic cycle and reactivation in the latent cycle. ICP0 transactivates gene expression by orchestrating an overall mitigation in host intrinsic/innate restrictions. How ICP0 coordinates its multiple active domains and its diverse protein-protein interactions is a key question in understanding the HSV-1 life cycle and pathogenesis. The present study focuses on delineating the regulatory effects of the SUMO-SIM interaction on ICP0 E3 ubiquitin ligase activity regarding PML II degradation. For the first time, we discovered the importance of multivalency in the PML II-ICP0 interaction network and report the involvement of different regulatory mechanisms in PML II recognition by ICP0 in HSV-1 infection.
INTRODUCTION
The infected cell protein 0 (ICP0) of herpes simplex virus 1 (HSV-1) is a unique viral regulator important for both the lytic and latent infection cycles of the virus. This is a multifunctional immediate early protein that mounts concerted counteractions to alleviate host intrinsic and innate restrictions (1). A main functional domain of ICP0 is its RING-type E3 ubiquitin ligase located in the second exon (2), by which ICP0 targets cellular proteins for proteasome-dependent degradation. Known ICP0 substrates include promyelocytic leukemia (PML) protein (3), speckled protein 100 (Sp100) (3), the catalytic subunit of DNA-dependent protein kinase (DNA-PKcs) (4), interferon inducible protein 16 (IFI16) (5), and RING finger protein 8 (RNF8) (6). A collection of evidence has shown that depletion of ICP0 E3 substrates, or sometimes their associated proteins, can improve HSV-1 viral replication in the absence of ICP0 (6–8), implying that these cellular substrates of ICP0 impose restrictions on HSV-1, which are maneuvered quickly by ICP0 in order to establish an effective viral replication.
One well-known ICP0 E3 substrate, PML, is the organizer protein for nuclear domains 10 (ND10s), aka PML nuclear bodies (PML-NBs), the dynamic nuclear structures involved in many important cell functions such as genome integrity, gene regulation, cell-cycle control, and antiviral responses (9–11). The PML protein has seven isoforms that differ in their C termini, resulting from alternative splicing of a single PML gene, whereas the shared N terminus has a tripartite motif (TRIM), three lysine residues that can be modified by small ubiquitin-like modifier (SUMO), and a SUMO-interaction motif (SIM) (12, 13). The RING domain in TRIM triggers self-tetramerization of PML proteins (14). Via self-tetramerization and the intermolecular SUMO-SIM interactions, various PML isoforms condensate to form a shell for ND10, whereas many other ND10 components are encircled as the inner core of ND10 (15, 16). Early in HSV-1 infection, ICP0 targets PML and another ND10 constituent, Sp100, for proteasome-dependent degradation (3). This leads to the disaggregation of ND10 and the release of viral repression imposed by ND10. Consequently, viral gene expression is promoted and viral replication is enhanced (11, 17).
Besides serving as an E3 ubiquitin ligase to target host restrictive factors, ICP0 also interacts with various cellular proteins (2) to modulate cell pathways. For example, ICP0 interacts with CoREST, a corepressor protein in the REST/CoREST/HDAC chromatin repressor complex, to dislodge HDAC from the complex and alter histone acetylation status on the viral genome. Consequently, the ICP0-CoREST interaction modulates the chromatin repression imposed on the viral genome and enhances viral expression (18, 19). A powerful feature of ICP0 is to interweave its E3 ligase activity with its protein-protein interactions, so that different functional domains coordinate to target multiple cellular restrictions for the purpose of orchestrating a robust infection. For example, mutations in the CoREST binding site have negative impacts on PML degradation, suggesting that ICP0 manipulates the ND10 restriction and chromatin repression in a cooperative fashion (20).
The coordination of ICP0 multiple functions becomes more complex with the discovery of SIMs scattering throughout the ICP0 sequence (21). SUMO is a member of the ubiquitin-like protein (UBL) superfamily that plays essential roles in a variety of important cellular functions, including nuclear structure, DNA repair, protein degradation, and cell immunity (22, 23). The multivalent SUMO-SIM interactions among the ND10 components provide a major molecular foundation for protein networking in ND10 (9, 15). The presence of SIMs in ICP0 allows ICP0 to directly interact with SUMOylated substrates such as PML, but it may also facilitate indirect protein networking with other ND10 components using PML as a bridge. Delineation of the relation between ICP0 sequence-specific interactions and SUMO-SIM interactions helps to understand the molecular basis of ICP0 functional cooperativity, which is a key stepping stone to demystifying the important role of ICP0 in HSV-1 lytic infection, as well as latency reactivation.
In a preceding report, we demonstrated that ICP0 can differentially interact with and degrade individual PML isoforms. We showed that PML isoform I was recognized as an ICP0 E3 substrate via a bipartite PML I-interacting domain flanking the RING finger domain, while the recognition of PML isoforms II, IV, and VI by ICP0 required a single SIM located at ICP0 residues 362 to 364 (SIM362-364) (24). Ubiquitin modification regulates not only protein stability, but also protein interaction and protein activity (25, 26). ICP0 differentially recognizing PML isoforms in ND10 may allow ICP0 to access other ND10 components, via SUMO-SIM interaction, in different manners. Because the original ND10 foci are where the virus eventually establishes replication compartments (27, 28), ICP0 may approach and harness ND10 proteins that are beneficial for viral replication. For example, CoREST is recruited to accumulate nearby ND10 and is found overlapping with viral replication compartments (28). To that end, delineating the molecular basis for ICP0 substrate differentiation becomes keystone to unlocking the mechanisms of early HSV-1 events abutting ND10. In the present study, we used PML II as the sample substrate to characterize the involvement of various ICP0 domains in SUMO-dependent substrate degradation. We investigated the specificity of various ICP0 SIMs by replacing them with SIMs from RNF4, a cellular SUMO-targeted ubiquitin ligase (STUbL) required to mediate the arsenic-induced proteasomal degradation of PML in the arsenic treatment of acute promyelocytic leukemia (29). RNF4 contains four consecutive SIMs with a higher affinity for poly-SUMO than that of mono-SUMO in vitro and has preferred targeting for SUMOylated PML (29). We found that a single SIM362-364 located in the center of ICP0 can be replaced by the four RNF4 SIMs, while using only two of the RNF4 SIMs was not effective at all. We also found that the three consecutive SIMs in the ICP0 C terminus can be replaced by the four RNF4 SIMs, whereas two of them can partially compensate, suggesting that SUMO-SIM interactions based in the central and C-terminal regions regulate PML II degradation via different mechanisms. More interestingly, disruption of the SUMO-SIM interaction between PML II and other ND10 components negatively impacted ICP0 recognition of these other ND10 components, whereas disruption of the ICP0 interaction with CoREST or ubiquitin-specific protease 7 (USP7), another important ICP0 binding partner (30), also affected the efficiency of PML II degradation, suggesting that substantial protein networking bridged by the SUMO-SIM interaction, as well as sequence-specific partner interactions, cooperatively regulate PML II degradation.
RESULTS
Two RNF4 SIMs are insufficient to replace a single ICP0 SIM362-364 in PML II degradation.
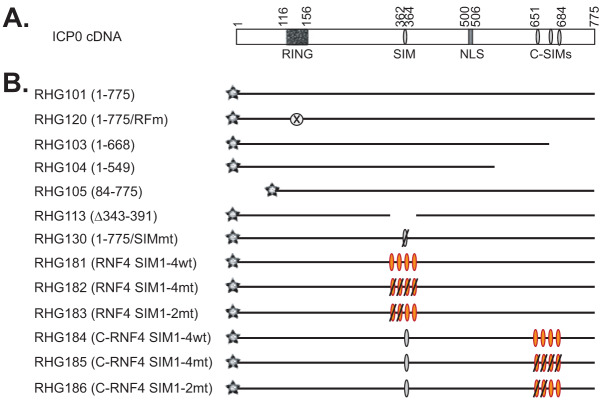
Previously, we reported that in HSV-1-infected HEp-2 cells, ICP0 requires the presence of SIM362-364 to interact with PML II and mediate PML II degradation (24). ICP0 has been defined as a STUbL due to this SUMO-dependency (21). To better understand how SUMO-interaction regulates ICP0 E3 ligase activity in recognizing PML II, we first asked whether SIM362-364 of ICP0 is virus specific. RNF4 is a cellular STUbL that triggers PML ubiquitination via its four consecutive SIMs interacting with the poly-SUMO chain of the SUMOylated PML (29). To examine whether the one SIM362-364 is equivalent to the four SIMs of RNF4, we replaced the ICP0 region 343 to 391 with RNF4 SIMs (wild-type or mutant form) and constructed recombinant viruses RHG181, 182, and 183, as shown in Fig. 1.
FIG 1.
Schematic diagram of recombinant viruses used in this study. (A) ICP0 cDNA and its main active domains. (B) Names of the recombinant viruses and their ICP0 gene structure. Gray star represents mCherry tag fused to the N terminus of all ICP0 constructs; gray oval represents ICP0 SIM; orange oval represents RNF4 SIM; an oval with a slash represents a mutated SIM; the crossed-out circle represents the mutated RING finger domain; and absence of line represents a deletion in ICP0 sequence.
In order to quantify the ICP0 ability to control the protein stability of PML isoforms, we have developed a half-life assay (24) in which we infect tetracycline-inducible cell lines stably expressing individual PML isoforms with ICP0 mutant viruses and then measure the effects of ICP0 mutations on the stability of a particular PML isoform (Fig. 2A). As previously described, tetracycline regulation allows us to stably introduce an exogenous cell cycle regulator, such as PML, without affecting cell growth during the stable line selection. In the meantime, the uniform expression of a stable line enables a quantitative analysis of ICP0 function in regulating PML stability (24).
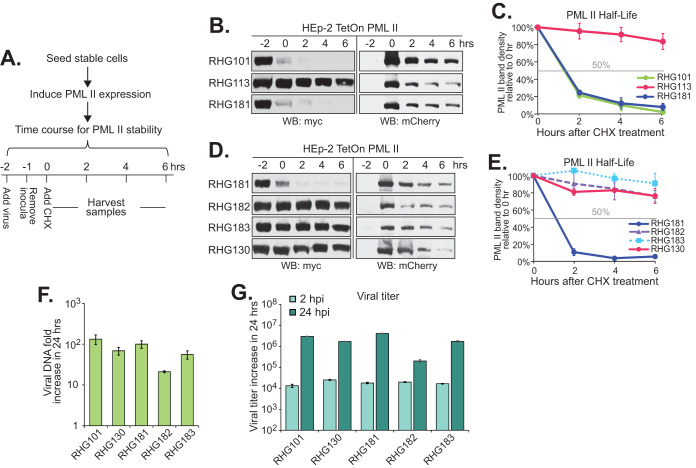
FIG 2.
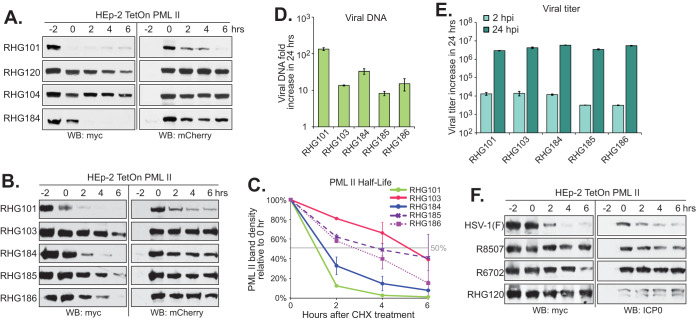
Requirement of all four RNF4 SIMs when replacing ICP0 SIM362-364 in PML II degradation. (A) Experimental design of the PML II half-life assay. HEp-2 TetOn cells stably expressing PML II were seeded and induced by doxycycline for 24 h. Cells were then infected with recombinant viruses at 10 PFU/cell. The time of virus addition was marked as −2 h. The time of cycloheximide addition was marked as 0 h. (B to E) Effects of ICP0 central SIM on the PML II half-life. Half-life assays for PML II were conducted with indicated viruses and myc-tagged PML II and mCherry-tagged ICP0 at different time points after cycloheximide treatment were detected by Western blotting (B and D). PML II band density from three (C) or two (E) independent experiments was quantified by Image J and plotted with the standard error to show the PML II half-life in different infections. (F and G) HEp2 TetOn cells expressing PML II were infected by the indicated viruses at 0.1 PFU/cell to measure growth properties. Viral DNA fold increase from 2 hpi to 24 hpi was measured with qPCR targeting ICP27 and 18S rDNA for viral DNA and total cell DNA, respectively, and calculated by 2−ΔΔCt using 2 hpi as the reference (F), whereas viral titer increase from 2 hpi to 24 hpi was measured in U2OS cells (G).
Consistent with our previous observations, when the cells were infected with RHG101 that contains the wild-type ICP0, myc-tagged PML II was quickly degraded after adding the virus, whereas ICP0 lacking residues 343 to 391 (RHG113) completely lost its ability to cause PML II degradation (Fig. 2B) (24). Interestingly, replacement of ICP0 residues 343 to 391 with the RNF4 residues 28 to 76 containing the four consecutive SIMs fully restored ICP0 capability for PML II degradation in the RHG181-infected cells (Fig. 2B and D). The restoration of ICP0 E3 ligase activity against PML II relied completely on the four RNF4 SIMs. When the hydrophobic residues in all four RNF4 SIMs were mutated, PML II degradation was again abolished in the RHG182-infected cells (Fig. 2D). Since the expression levels of mCherry-tagged mutant ICP0 were comparable in cells infected with the different recombinant viruses (Fig. 2B and D, right), we concluded that ICP0 E3 activity required the presence of SIM(s) in the central region of ICP0 to recognize PML II, but the source of SIM had no specificity.
Tatham et al. showed that multiple SUMO-SIM interactions from RNF4 were needed to ubiquitinate PML in vitro (29). However, SIM362-364 is the only SIM present in the central region of ICP0. To investigate whether all four RNF4 SIMs were needed to rescue the loss of SIM362-364 in ICP0, we mutated the hydrophobic residues in the first two SIMs and maintained RNF4 SIM-3 and SIM-4 in their original sequences, and constructed recombinant virus RHG183. As shown in Fig. 2D, the myc-tagged PML II in RHG183-infected cells was stable throughout the course of the half-life assay, similar to that of RHG130, a recombinant virus containing mutated SIM362-364 in ICP0 (24). Quantitation of the PML II band density showed that while RHG101 and RHG181 infection massively destabilized PML II to a half-life of ∼1 h, the half-life of PML II in both RHG182- and RHG183-infected cells was longer than 6 h, similar to that of ICP0 with mutated or deleted SIM362-364 (Fig. 2C and E). Therefore, we concluded that two RNF4 SIMs were not sufficient to compensate for the loss of a single SIM362-364 in ICP0. Although SIM362-364 had no sequence specificity for determining the interaction between ICP0 and PML II, sequence surrounding SIM362-364 in ICP0 facilitated the single SUMO-SIM interaction, likely by a secondary mechanism to substitute for the requirement of multiple SIMs.
We further examined the effects of SIM replacement on the replication of recombinant viruses. Since ICP0 lacking SIM362-364 is capable of degrading PML I (24), and likely many other SUMO-independent substrates, only minor changes were observed in replication within 24 h postinfection (hpi), at both viral DNA and viral production levels (Fig. 2F and G).
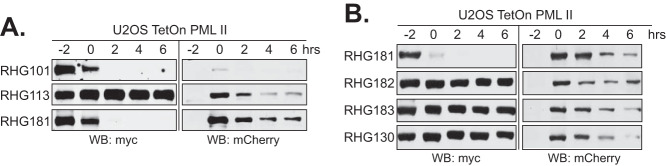
SIM362-364 in the central region of ICP0 regulates PML II degradation in a cell-type-independent manner.
ICP0 functions to counteract multiple fronts of the host intrinsic/innate antiviral defense (2, 11, 31). In nonpermissive cells, ICP0 is essential for viral replication at low multiplicity of infection (MOI) but is dispensable at high MOI. Likely the preexisting antiviral factors that need to be disabled by ICP0 can be saturated by a higher dose of viral input (1, 2). U2OS cells are known to be permissive to the ICP0-null virus for its lack of certain restrictive factors (32–34). In a preceding report, we showed that SIM362-364 is also required for PML II degradation in the permissive U2OS cells (24). To further evaluate whether HEp-2 and U2OS cells share the same broader regulations for PML II degradation in HSV-1 infection, we conducted the same half-life assays for viruses containing RNF SIM4 in the place of SIM362-364 in U2OS cells stably expressing PML II. Consistent with what we saw in HEp-2 TetOn PML II cells, results in Fig. 3A show that replacement of SIM362-364 by four RNF4 SIMs also restored PML II degradation in the RHG181-infected U2OS cells. Mutations in all four RNF SIMs completely abolished PML II degradation, whereas keeping two functional RNF4 SIMs was not sufficient to rescue the PML II degradation (Fig. 3B). From this we conclude that the primary regulatory mechanism of PML II degradation in HSV-1 infection is cell-type independent.
FIG 3.
Regulation of RNF SIMs on the PML II degradation is a cell-type-independent phenomenon. U2OS TetOn cells stably expressing PML II were subjected to the PML II half-life assay with the indicated viruses (A and B) as described above.
ICP0 C terminus is involved in the SIM362-364-dependent substrate degradation.
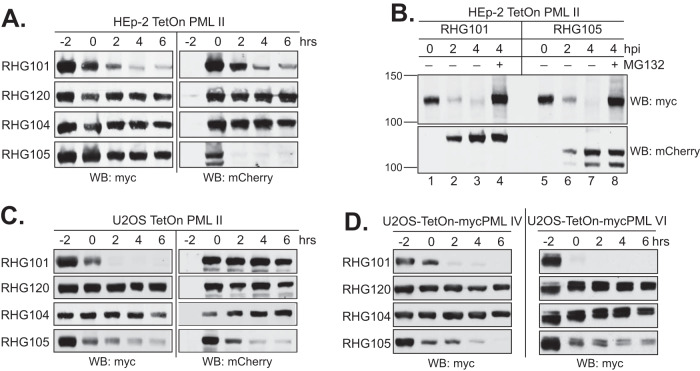
To further examine whether ICP0 domains other than the central region of ICP0 are also involved in PML II degradation, we infected cells with recombinant viruses lacking either the N-terminal residues upstream of the RING domain (RHG105) or the entire C-terminal region (RHG104) (Fig. 1). As shown in Fig. 4A, PML II was fairly stable in both RHG104- and RHG105-infected cells, similar to that for the RHG120 virus, which contains C116G/C156A substitutions and thereby an inactive RING domain (Fig. 4A). Interestingly, ICP0 deleted of N-terminal residues 1 to 83 was unstable by itself upon cycloheximide treatment, for which degradation of PML II did not progress due to the disappearance of ICP0. To further assess whether the ICP0 N terminus has a direct role in PML II degradation or the quick loss of ICP0 lacking the N terminus had an indirect impact on PML II degradation, we examined the PML II level in RHG105 infection without the cycloheximide treatment. As shown in Fig. 4B, PML II in cells infected with RHG105 had a quick decrease in the steady-state level within the first 4 h after infection, representing a degradation pace similar to that of RHG101. We also showed that the decrease of PML II level was completely blocked when proteasome inhibitor MG132 was added to the RHG105 infection, consistent with what happened within RHG101 infection (lanes 4 and 8).
FIG 4.
ICP0 C terminus, but not the N terminus, is necessary for the degradation of PML II, PML IV, and PML VI in both HEp-2 and U2OS cells. (A and C) Effects of ICP0 N and C termini on the PML II half-life in HEp-2 TetOn cells (A) or U2OS TetOn cells (C). (B) Degradation of PML II by ICP0 lacking the N terminus in the absence of cycloheximide. HEp-2 TetOn cells expressing PML II were induced and infected by the indicated viruses in the presence or absence of MG132. At 0, 2, and 4 hpi, total cell lysates were harvested and myc-tagged PML II and mCherry-tagged ICP0 were detected by Western blotting. (D) Effects of ICP0 N and C termini on the PML IV (left) and PML VI (right) half-lives in U2OS cells.
We further examined the N- and C-terminal effects on PML II degradation in the permissive U2OS cells. In these cells, we found the ICP0 C terminus is also important to PML II degradation, whereas the deletion of the ICP0 N terminus had a lesser effect (Fig. 4C, left). Interestingly, ICP0 lacking the N-terminal residues 1 to 83 was more stable in U2OS cells compared to HEp-2 cells (Fig. 4A and C, right), implying that the stability of ICP0 is correlated with its functionality. Therefore, when the target(s) is missing and the function is dispensable in the permissive U2OS cells, ICP0 is more stable.
Previously, we have shown that the degradation of PML isoforms IV and VI in HSV-1 infection also required the presence of SIM362-364 (24). To examine whether these two PML isoforms follow the same regulatory mechanism as that of PML II, we infected U2OS TetOn cells expressing PML IV or PML VI with RHG104 and RHG105. Similar to that of PML II (Fig. 4C), degradation of PML IV and PML VI also relied on the presence of the ICP0 C terminus but not so much on the ICP0 N terminus (Fig. 4D). Taken together, we conclude that the regulatory mechanism of PML II degradation in HSV-1 infection is shared by other ICP0 substrates requiring SIM362-364.
ICP0 C terminus regulates PML II degradation by multiple mechanisms.
There are three SIMs located in the ICP0 C terminus, at residues 651 to 654, 667 to 670, and 681 to 684 (21). ICP0 truncated at residue 668 (RHG103) still has one of the C-terminal SIMs remaining, whereas truncation at residue 549 (RHG104) has all C-terminal SIMs deleted (24). Consistent with the previous report, PML II degradation triggered by ICP0 was largely abolished in both RHG104- or RHG103-infected cells (Fig. 5A and B). To examine whether the three consecutive SIMs in the ICP0 C terminus function similarly compared to SIM362-364, we removed ICP0 residues 651 to 684, replaced them with the RNF4 residues 36 to 70 that encompass the four RNF4 SIMs, and constructed recombinant virus RHG184 (Fig. 1). Figure 5A shows that the restoration of ICP0 C-terminal sequences 685 to 775 and the insertion of RNF4 SIMs in RHG184 resuscitated ICP0’s ability to mediate PML II degradation. Figure 5C demonstrates that the half-life of PML II in RHG184-infected cells was slightly longer than that of RHG101, but it was still less than 2 h, suggesting a substantial restoration of the PML II degradation by the RNF4 SIMs and ICP0 C terminus. To further evaluate the roles of the RNF4 SIMs and ICP0 downstream sequences in the regulation of PML II degradation, we mutated all four RNF4 SIMs or only the first two RNF4 SIMs and constructed recombinant viruses RHG185 and RHG186, respectively. Different from what was observed in Fig. 2D and E, mutations in four or two of the RNF4 SIMs did not abolish PML II degradation and PML II half-life was delayed to 4 h and 3 h in RHG185- and RHG186-infected cells, respectively (Fig. 5B and C). Therefore, we conclude that the presence of multiple SIMs in the ICP0 C terminus plays a secondary role compared to SIM362-364 and additively facilitates the degradation of PML II. Consistent with what we observed in Fig. 2F and G, mutations in the C-terminal SIMs of ICP0 did not affect the overall replication properties of the recombinant viruses (Fig. 5D and E).
FIG 5.
Involvement of the ICP0 C-terminal active domains in PML II degradation. (A and B) PML II half-life assays in HEp-2 TetOn PML II cells infected by the indicated viruses as described above. (C) PML II band density from two independent experiments was quantified by ImageJ and plotted with standard error. (D and E) Viral growth properties measured by DNA fold increase (D) and titer increase (E) from 2 hpi to 24 hpi as described above. (F) Effects of mutations in the CoREST-binding site or USP7-binding site on the PML II half-life.
The ICP0 C terminus is a highly active region, containing the CoREST binding site located in residues 671 to 673 (20) and the USP7 binding site located in residues 620 to 624 (35) in the vicinity of C-terminal SIMs. ICP0-CoREST interaction counteracts the chromatin repression imposed by the REST/CoREST/HDACs repressor complex (18, 19), whereas ICP0-USP7 interaction has been suggested to reciprocally regulate the stability for the paired ICP0 E3 ligase and USP7 deubiquitinase (36).
To further understand the role of ICP0 C-terminal domains in the coordination with its E3 ubiquitin ligase activity, we conducted a PML II half-life assay with recombinant virus R8507, which has the D671A/E673A substitutions that knock out the CoREST binding (20), and recombinant virus R6702, which has the K620I mutation that knocks out the USP7 binding (37). Results showed that both R8507 and R6702 were defective in PML II degradation (Fig. 5F), suggesting that ICP0 interactions with other functional partners can heavily affect how ICP0 E3 ubiquitin ligase targets PML II in HSV-1 infection.
Disrupting the SUMO-SIM interaction of PML II abolishes its degradation in HSV-1 infection but not its ND10 colocalization.
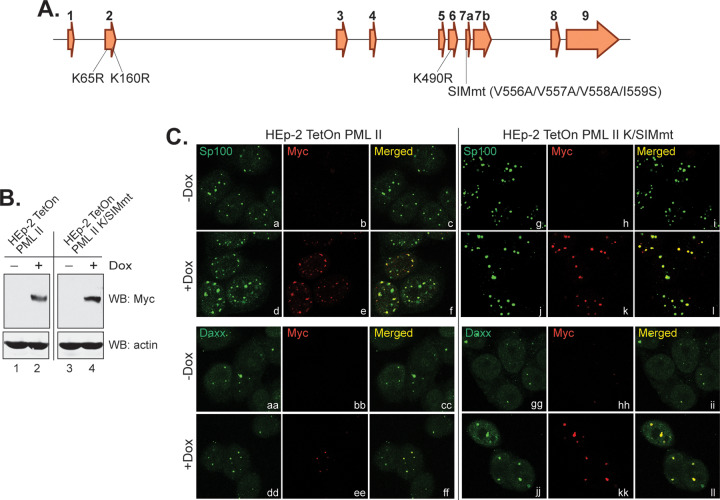
Interactions between the SUMOylated proteins and SIM-containing proteins provide molecular foundations for the ND10 organization (9, 38). As the organizer protein, all nuclear PML isoforms contain three SUMOylation sites at lysine residues 65, 160, and 490 (13) and one SIM at residues 556 to 559 (39). To further understand the effects of protein networking and ND10 organization on PML II degradation, we mutated both SIM and SUMOylation sites in PML II (Fig. 6A) and constructed a tetracycline-inducible cell line HEp-2 TetOn-PML II K/SIMmt. Similar to the wild type PML II stable line (24) (Fig. 6B, lanes 1 and 2), PML II K/SIMmt expression is also tightly controlled by the tetracycline repressor and is highly induced upon doxycycline treatment (Fig. 6B, lanes 3 and 4).
FIG 6.
Expression and subcellular localization of a PML II mutant containing substitutions in SUMOylation sites and the SUMO-interaction motif. (A) Schematic diagram of PML II gene structure, with exon numbers listed on the top of each exon and positions of amino acid substitutions marked below the exons. (B) Induction of PML II K/SIM mutant compared to wild-type PML II in HEp-2 TetOn cells. (C) Colocalization of myc-tagged wild-type PML II or PML II K/SIMmt with Sp100 and Daxx in the ND10 nuclear bodies in HEp-2 TetOn cells.
We further examined the subcellular localization of PML II K/SIMmt and found that PML II K/SIMmt is competent to colocalize to ND10s represented by either Sp100 or Daxx (Fig. 6C, j to l and jj to ll), the same as that of the ectopic wild-type PML II (Fig. 6C, d to f and dd to ff). These results indicated that blocking SUMOylation and SUMO-interaction in PML II did not prevent the mutant PML II from associating with other ND10 components, nor did it drastically change the size, number, and shape of ND10s. It is known that the coiled-coil domain of TRIM located within residues 229 to 323 of all PML isoforms is required for the self-oligomerization of PML (13). Likely the presence of a functional coiled-coil domain in the PML II K/SIMmt mutant allows it to multimerize with the endogenous PML isoforms, triggering the aggregation of PML II K/SIMmt to ND10 as shown in Fig. 6C.
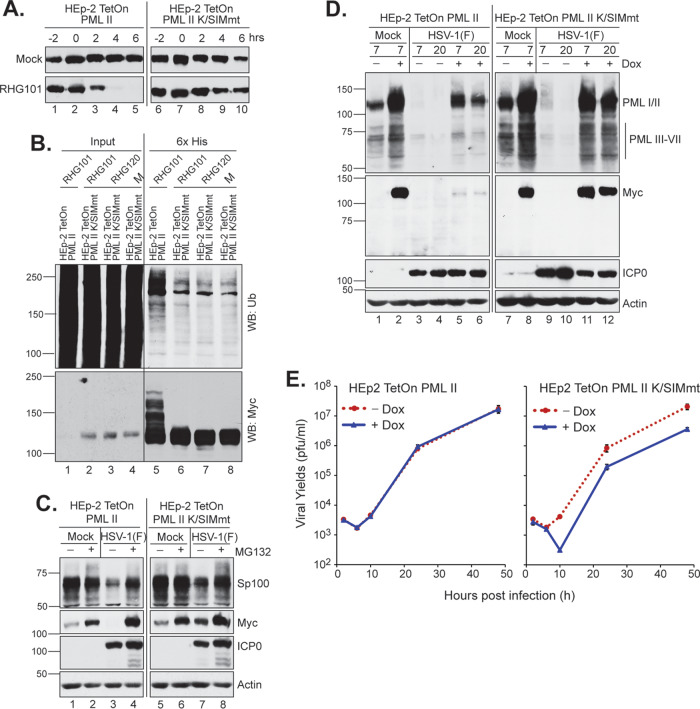
To examine whether the PML II K/SIMmt mutant can be degraded by ICP0 in HSV-1 infection, we performed a half-life assay in the HEp2-TetOn PML II K/SIMmt cell line. Figure 7A shows that while the wild-type PML II was readily degraded (lanes 2 to 5) by the wild-type ICP0 in RHG101 infection, PML II K/SIMmt was resistant to such degradation (lanes 7 to 10). We further confirmed this conclusion with an in vivo ubiquitination assay. When we built the expression plasmids of myc-tagged PML II and PML II K/SIMmt, we used pcDNA4/TO/LacZ plasmid (Invitrogen) as the vector and replaced the LacZ gene with the myc-PML II cassette. The pcDNA4/TO/LacZ plasmid has a 6×His tag fused to the N terminus of LacZ, which was retained in frame with PML II or PML II K/SIMmt. With the 6×His tag, we performed Ni-NTA pulldown under denaturing conditions and examined the ubiquitination level of PML II and PML II K/SIMmt. Results showed that while ubiquitinated PML II of high molecular weight was readily detectable in cells infected by RHG101 (Fig. 7B, lane 5), ICP0-mediated ubiquitination was not observed for PML II K/SIMmt compared to the negative controls of mock- or RHG120-infected cells (Fig. 7B, lanes 6 to 8). Therefore, SUMOylation is essential in the PML II interaction with ICP0 for its degradation. ND10 localization of PML II K/SIMmt is not sufficient to facilitate the substrate recognition by ICP0.
FIG 7.
Effects of K/SIM mutations on the degradation of PML II and PML II-associated proteins. (A) Half-life assays for wild-type PML II and PML II K/SIMmt in RHG101-infected cells as described above. (B) K/SIM mutations block PML II ubiquitination in RHG101-infected cells. HEp-2 TetOn cells expressing wild-type PML II or PML II K/SIMmt were induced, infected, and harvested for Ni-NTA pulldown under denaturing conditions. PML II precipitates were then probed with anti-Ub (upper) and anti-myc (lower) antibodies. (C and D) Effects of PML II K/SIM mutant on the degradation of Sp100 (C) and PML isoforms III-VII (D) in HEp-2 TetOn cells infected by HSV-1(F) at 5 PFU/cell. (E) HEp-2 TetOn cells expressing wild-type PML II (left) or PML II K/SIMmt (right) were infected by HSV-1(F) at 0.1 PFU/cell in the presence or absence of 1 μg/ml of doxycycline. At 2, 6, 10, 24, and 48 hpi, triplicate infected cells were harvested and titrated on U2OS cells. Titers with the standard error were plotted by Microsoft Excel.
PML II K/SIMmt has a negative impact on viral replication.
Since PML II K/SIMmt is colocalized to the endogenous ND10 (Fig. 6C), we postulated that the presence of PML II K/SIMmt in PML oligomerization may affect the self-interaction among PML isoforms and consequently the interaction between PML and other ND10 components. To test this hypothesis, we first evaluated the degradation of endogenous Sp100 in PML II wild-type versus mutant cells infected by HSV-1(F). Figure 7C shows a mild delay in Sp100 degradation in cells expressing PML II K/SIMmt, in the absence of proteasome inhibitor MG132, compared to that in the wild-type PML II-expressing cells (lanes 3 and 7). We further quantitated the Sp100 band and normalized it against the actin band by Image J software. Results showed that using mock infection as the reference, HSV-1 infection caused a 42% loss of Sp100 in the PML II K/SIMmt cells (Fig. 7C, lane 7) and a 54% loss in the wild-type PML II cells (Fig. 7C, lane 3) at 12 h postinfection. We also examined the dominant-negative effect of mutant PML II on the endogenous PML degradation. PML isoforms I and II are similar in size, so the overexpression of exogenous PML II (wild-type or mutant) will obscure the amount of endogenous PML I and II when probed by an antibody recognizing all PML isoforms. The size of PML isoforms III to VII ranges from 55 to 70 kDa. We clearly detected a cluster of bands in that range by the PML antibody but not by the anti-myc antibody, suggesting these bands are the various forms of endogenous PML III to VII (Fig. 7D). All these forms were upregulated by the doxycycline induction, implying a coordinated stoichiometric balance among PML isoforms. These isoforms were mostly degraded by HSV-1 infection in PML II-expressing cells, but not so much in the PML II K/SIMmt-expressing cells. We again quantitated the cluster of bands and normalized them against actin. Compared to the mock infection (Fig. 7D, lanes 2 and 8), HSV-1 infection caused a 69.1% and 69.5% loss of PML III to VII in wild-type PML II-expressing cells at 7 and 20 hpi (Fig. 7D, lanes 5 and 6), but only a 49.4% and 50.0% loss in PML K/SIMmt-expressing cells (Fig. 7D, lanes 11 and 12). The mild delay in the degradation of endogenous Sp100 and PML in PML II K/SIMmt cells also showed a significant impact on HSV-1 viral production. Shown in Fig. 7E, while HSV-1(F) showed the same pace of growth in the wild-type PML II-expressing cells, with or without the doxycycline induction of exogenous PML II, the growth of HSV-1(F) in cells induced for PML II K/SIMmt expression had a 13-fold decrease compared to the uninduced cells at 10 hpi and an ∼5-fold decrease at 24 and 48 hpi. The larger delay in viral replication observed at 10 hpi indicated that the dominant-negative effect of PML II K/SIMmt had more impacts on the immediate early events and the onset of DNA replication.
DISCUSSION
ICP0 is a key viral regulator in HSV-1 infection that orchestrates counteractions against multiple intrinsic/innate antiviral defenses in order to enhance the overall viral replication and productivity. The E3 ubiquitin ligase located in the RING finger domain of ICP0 is the most prominent activity of this protein. The ICP0 E3 ligase seems to be tightly controlled by the proximal or distal elements encompassing many active domains of ICP0, and possibly also by other cellular/viral factors (2). Due to the pivotal position ICP0 serves in both the lytic and latent cycles of HSV-1 infection (1), delineating how ICP0 E3 is regulated and determining the consequences of such regulation is essential for understanding the HSV-1 infection and pathogenesis. We previously reported that two very similar substrates, PML I and PML II, are differentially recognized by the ICP0 E3 ubiquitin ligase. While SIM362-364 is essential for PML II to be recognized and degraded, it is completely dispensable for the PML I degradation (24). The present study focuses on understanding the regulation of SUMO-SIM interaction on the ICP0 E3 ligase activity.
The SUMO moiety provides several binding interfaces that can interact with proteins differently. Typical SIMs containing a short stretch of hydrophobic amino acids followed by acidic amino acids are defined as class I SUMO interaction motifs (40). ICP0 SIM362-364 and the four RNF4 SIMs all have the conserved class I sequence. However, when we replaced the single SIM362-364 with four consecutive SIMs from RNF4, we found that the two active RNF4 SIMs are insufficient to compensate for the loss of a single SIM362-364 (Fig. 2). In the ICP0 region 343 to 391 surrounding SIM362-364, there are 14 proline residues and some of them are assembled into the PxxP motif, the consensus sequence recognized by the SH3 domain (41, 42). Both SH3-PxxP interaction and SUMO-SIM interaction can provide a molecular foundation to organize protein condensates through liquid-liquid phase separation (43). Our previous results showing that three proline-rich ND10-fusion segments in the central region of ICP0 control the access of ICP0 from docking at the ND10 surface to fusing into the core of ND10 (44) strongly suggest the involvement of these PxxP motifs in the interaction between ICP0 and ND10 components. Likely, the presence of SIM362-364 alongside the proximal tandem PxxP motifs enhances the multivalent interactions required for the ICP0-mediated PML II recognition and degradation. When the region of 343 to 391 is replaced by RNF4 SIMs, which lack the PxxP motifs, the PMLII-ICP0 interaction has to rely solely on SUMO-SIM interaction and the presence of more than two SIMs becomes required to maintain the minimum multivalency. Since mutations in the three hydrophobic residues in SIM362-364 completely block PML II degradation (Fig. 2), how PxxP motifs in the vicinity of SIM362-364 work to influence ICP0 E3 activity requires further investigation.
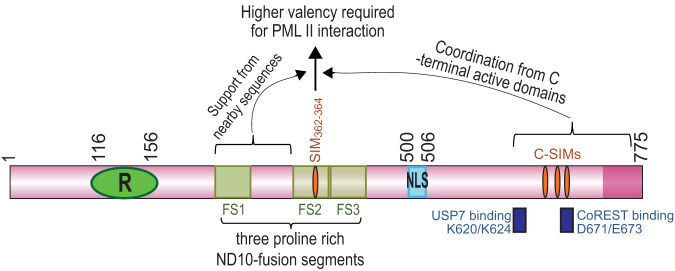
In a preceding report, we showed that ICP0 sequences nearby residues 343 to 391 are also important for PML II degradation (24). The presence of residues 343 to 391 in the central region encompassing ICP0 residues 244 to 474 is sufficient to allow PML II to bind with ICP0 but not for for PML II degradation, whereas partial restoration of sequences surrounding residues 343 to 391 can gradually reconstitute the PML II degradation (24). In the present study, we found that the ICP0 C terminus is also important for PML II degradation. In contrary to region 343 to 391, RNF4 SIMs replacing the ICP0 C-terminal SIMs showed a correlation between the number of RNF4 SIMs and the extent of PML II degradation, with two RNF4 SIMs partially rescuing and four RNF4 SIMs fully reconstituting the PML II degradation (Fig. 5). In addition, mutations in the C-terminal CoREST- and USP7-binding sites also greatly mitigated the PML II degradation caused by ICP0 E3 (Fig. 5F). Taking all these observations together, we conclude that different biochemical mechanisms govern the interaction between PML II and the various ICP0 active domains. The interaction of PML II with the ICP0 central residues 343 to 391 has a pivotal role in PML II degradation, and has a high threshold of multivalency required to maintain a strong interaction. Sequences surrounding residues 343 to 391 progressively improve the ability of this region to degrade PML II. In the presence of a complete central region, the C-terminal SIMs of ICP0 and sequences in the vicinity of these SIMs orchestrate the enhancement of PML II degradation. In Fig. 8, the two regions proximal and distal to residues 343 to 391 are illustrated to show their auxiliary roles in the PML II degradation, but it remains unclear whether this is due to an overall structural support or the involvement of additional protein-protein interactions. In silico protein modeling shows unstructured property in both the ICP0 central and C-terminal regions (unpublished data). Moreover, both the USP7-binding site and the CoREST-binding site are in the vicinity of the ICP0 C-terminal SIMs (Fig. 8). Understanding how these flexible regions of ICP0, together with the complex protein interaction network, coordinately affect the multivalent primary interactions between PML II and ICP0 residues 343 to 391 will be key to understanding the ICP0 cooperativity in HSV-1 infection.
FIG 8.
Coordination among ICP0 active domains in its recognition and degradation of PML II. The pink rod represents the 775-amino acid ICP0 cDNA; green oval with an R represents the RING finger domain located at residues 116 to 156; yellow squares represent the three proline-rich ND10-fusion segments (FS1, FS2, and FS3) identified in a previous report (44); orange ovals represent SIMs in the central and C-terminal regions of ICP0, with SIM362-364 located within FS2; blue rectangle with NLS inside represents nuclear localization sequence at residues 500 to 506; dark blue rectangles beneath ICP0 indicate the positions of USP7- and CoREST-binding sites.
SUMO-SIM interaction is a critical molecular foundation that supports protein condensation in the ND10 nuclear bodies. To understand whether ICP0 interacting with PML bridges additional interactions among ND10 components via the interwoven SUMO-SIM interaction, we constructed a PML II mutant that is inactive in both SUMOylation sites and the SIM. Mutations in these sites did not substantially change ND10 formation (Fig. 6). Likely, the intact TRIM domain in the mutant PML II is sufficient for self-oligomerization and draws it to ND10s via interaction with endogenous PML. The inability of the PML II mutant to undergo SUMO interaction completely blocked its degradation in HSV-1 infection, consistent with the previous result that mutation in SIM362-364 abolishes the degradation of wild-type PML II (24). Interestingly, this inability for SUMO interaction by the mutant PML II mildly delayed the degradation of endogenous PML and Sp100 (Fig. 7C and D), and subsequently the viral replication of HSV-1(F) (Fig. 7E). These results suggest that SUMO interaction among ND10 components affects how these individual components are recognized by ICP0. The dominant-negative effects of PML II K/SIMmt are less obvious in RHG101 infection (data not shown) than that in the prototype strain HSV-1(F) infection (Fig. 7), suggesting that the ICP0 ability to recognize ND10 components may have additional unknown regulations. It would be interesting to determine the coordination of ND10 organization in regulating the ICP0 functionality and HSV-1 replication, especially in the context of neuronal infection and immune surveillance.
MATERIALS AND METHODS
Cells and viruses.
HEp-2 TetOn cells were maintained in Dulbecco’s modified Eagle medium (DMEM) (Gibco) supplemented with 10% fetal bovine serum (FBS) (Invitrogen) and 6 μg/ml blasticidin (Invitrogen). HEp-2 TetOn cells stably expressing PML II were maintained in DMEM medium supplemented with 10% FBS plus 6 μg/ml blasticidin and 100 μg/ml zeocin (Invitrogen). U2OS TetOn cells expressing PML II, IV, or VI were maintained in McCoy 5A medium (Gibco) supplemented with 10% FBS plus 6 μg/ml blasticidin and 100 μg/ml zeocin (Invitrogen). Recombinant viruses RHG101, RHG120, RHG103, RHG104, RHG105, RHG113, and RHG130 have been previously described (24, 45). Prototype HSV-1(F) and recombinant viruses R8507 and R6702 are generous gifts from Bernard Roizman (University of Chicago) (20, 37).
Construction of chimeric viruses containing RNF4 SIMs in the central region of ICP0.
In order to fuse RNF4 SIMs into the center of ICP0, BglII and EcoRV sites were inserted in between Arg342 and Ser392 of ICP0 by a two-step jump strand PCR. Upstream primer pair 5′-GCAGGATCCGCAGACTACGTACCGCCCGC-3′ and 5′-TCCCGACGCGGCCGCGGAGATATCAGATCTCCCTCGGCCGCCCCGCCTC-3′, and downstream primer pair 5′-GAGGCGGGGCGGCCGAGGGAGATCTGATATCTCCGCGGCCGCGTCGGGA-3′ and 5′-GCAGTCGACTTACCCGGGCCCACCCTGGCCGCG-3′, were used in the first-step PCR to generate ICP0 fragments upstream of Arg342 and downstream of Ser392, with overlapping sequences containing BglII and EcoRV sites. The two PCR products were then annealed together to serve as the template for the second round PCR by the upstream primer 5′-GCAGGATCCGCAGACTACGTACCGCCCGC-3′ and downstream primer 5′-GCAGTCGACTTACCCGGGCCCACCCTGGCCGCG-3′. The resulting PCR product with the 343 to 391 deletion and BglII/EcoRV insertion was then digested with NruI and MluI to swap the NruI-MluI fragment of wild-type ICP0 into plasmid pHG101. The resulting plasmid was named pBKS-ICP0(1-342/BglII/EcoRV/392-775). Primers 5′-CGAGATCTCCTTGGAAGC-3′ and 5′-CGGATATCTGGTCTTCTTCTTTCGTC-3′ were used to PCR amplify the RNF4 residues 28 to 76. The PCR product was digested with BglII and EcoRV and inserted in between the BglII and EcoRV sites of plasmid pBKS-ICP0(1-342/BglII/EcoRV/392-775) to generate pHG181.
To mutate all four of the RNF4 SIMs, overlapping upstream primer pair 5′-GAGTCGCATCAGCCGCCACAGGCTCTAAAGATTCACAAGTGGCGTCCGCAGCTTCATCTC-3′ and 5′-CCGAGATCTCCTTGGAAGCAGAACCCGCAGAAGCCGCGGAAACTGCTGGAGATGAAGCTGCGGACGCC-3′, and overlapping downstream primer pair 5′-ACTTGTGAATCTTTAGAGCCTGTGGCGGCTGATGCGACTCACAATGACTC-3′ and 5′-CGGATATCCGCAGCCGCAGCAGAGTCATTGTGAGTCGCATCAGCCGCCACA-3′, were annealed by heating to 95°C for 5 min and then cooled down slowly. The annealed primers were then filled in by Klenow and mixed together to serve as the template for PCR amplification with primers 5′-CGAGATCTCCTTGGAAGC-3′ and 5′-CGGATATCTGGTCTTCTTCTTTCGTC -3′. The PCR product was digested with BglII and EcoRV and inserted in between the BglII and EcoRV sites of plasmid pBKS-ICP0(1-342/BglII/EcoRV/392-775) to generate pHG182.
To mutate the first 2 RNF4 SIMs, overlapping upstream primer pair 5′-CCGAGATCTCCTTGGAAGCAGAACCCGCAGAAGCCGCGGAAACTGCTGGAGATGAAGCTGCGGACGCC-3′ and 5′-GAGTCAGATC AACCACCACAGGCTCTAAAGATTCACAAGTGGCGTCCGCAGCTTCATCTC-3′ were annealed and filled in by Klenow as described above. Primers 5′-ACTTGTGAATCTTTAGAGCC-3′ and 5′-CGGATATCTGGTCTTCTTCTTTCGTC -3′ were used to PCR amplify the downstream fragment and then mixed with Klenow-filled upstream fragment to serve as the template for the second phase of jump strand PCR. The PCR product was digested with BglII and EcoRV and inserted in between the BglII and EcoRV sites of plasmid pBKS-ICP0(1-342/BglII/EcoRV/392-775) to generate pHG183.
The mutant ICP0 gene cassettes in pHG181, pHG182, and pHG183 were digested with PmeI and XbaI, inserted into the pKO5 plasmid, and electroporated into E. coli RR1 strain harboring an ICP0-null bacterial artificial chromosome (BAC) (18). DNA extracted from each recombinant BAC clone was transfected into U2OS cells and generated recombinant viruses RHG181, RHG182, and RHG183. Recombinant viruses were plaque purified at least three times in U2OS cells. Viral DNAs were isolated, and the existence of mutations in both copies of ICP0 was verified by Southern blotting and sequencing.
Construction of chimeric viruses containing RNF4 SIMs in the C terminus of ICP0.
In order to fuse RNF4 SIMs into the C-terminal region of ICP0, SpeI and EcoRI sites were inserted in between Ser650 and Asp685 of ICP0 by the two-step jump strand PCR. Upstream primer pair 5′-TCCCCCCAGTCCACGCGTCC-3′ and 5′-CATGTTTCCCGTCTGGTCGAATTCACTAGTGCTAGAGACCCCCGAGAT-3′, and downstream primer pair 5′-ATCTCGGGGGTCTCTAGCACTAGTGAATTCGACCAGACGGGAAACATG-3′ and 5′-TATTGTTTTCCCTCGTCCCG-3′, were used in the first-step PCR to generate ICP0 fragments upstream of Ser650 and downstream of Asp685, with overlapping sequences containing SpeI and EcoRI sites. The two PCR products were then annealed together to serve as the template for the second round PCR by the upstream primer 5′-TCCCCCCAGTCCACGCGTCC-3′ and the downstream primer 5′-TATTGTTTTCCCTCGTCCCG-3′. The resulting PCR product with the 651 to 684 deletion and SpeI/EcoRI insertion was then digested with MluI and SalI to swap the MluI-SalI fragment of wild-type ICP0 into plasmid pHG101. The resulting plasmid was named pBKS-ICP0(1-650/SpeI/EcoRI/685-775). Primers 5′-CCCACTAGTATAGAACTCGTGGAAACT-3′ and 5′-CCCGAATTCAACAATCACAACAGAGTC-3′ were used to PCR amplify the RNF4 residues 36 to 70. The PCR product was digested with SpeI and EcoRI and inserted in between the SpeI and EcoRI sites of plasmid pBKS-ICP0(1-650/SpeI/EcoRI/685-775) to generate pHG184. pHG182 and pHG183 were used as the template to PCR amplify the RNF4 SIM1-4mt and RNF4 SIM1-2mt with primers 5′-CCCACTAGTGCAGAAGCCGCGGAAACT-3′ and 5′-CCCGAATTCCGCAGCCGCAGCAGAGTCATTG-3′. The PCR products were digested with SpeI and EcoR I and inserted in between the SpeI and EcoRI sites of plasmid pBKS-ICP0(1-650/SpeI/EcoRI/685-775), which generated pHG185 and pHG186 plasmids, respectively.
The mutant ICP0 gene cassettes in pHG184, pHG185, and pHG186 were recombined into ICP0-null BAC and packaged into recombinant viruses RHG184, RHG185, and RHG186 as described above.
Construction of stable cell lines.
To construct an HEp-2 TetOn cell line stably expressing PML II K/SIM mutant, primer pairs 5′-CAATGCCAGGCGGAAGCCCGGTGCCCGAAGCTGCTGCC-3′ and 5′-GGCAGCAGCTTCGGGCACCGGGCTTCCGCCTGGCATTG-3′ for K65R substitution, 5′-GCACACCAGTGGTTCCTCCGGCACGAGGCCCGGCCCCTAG-3′ and 5′-CTAGGGGCCGGGCCTCGTGCCGGAGGAACCACTGGTGTGC-3′ for K160R substitution, 5′-TGCCCCAGGAAGGTCATCCGGATGGAGTCTGAGGAGGGG-3′ and 5′-CCCCTCCTCAGACTCCATCCGGATGACCTTCCTGGGGCA-3′ for K490R substitution, and 5′-GAGGCAGAGGAACGCGCTGCGGCGAGCAGCAGCTCGGAAGAC-3′ and 5′-GTCTTCCGAGCTGCTGCTCGCCGCAGCGCGTTCCTCTGCCTC-3′ for V556A/V557A/V558A/I559S substitutions were applied to plasmid pCDNA4/To/Myc-PML II sequentially. After 4 rounds of PCR mutagenesis by QuikChange II site-directed mutagenesis kit (Agilent Technologies), the final plasmid was named pcDNA4/TO/Myc-PML II K/SIM mut. The presence of mutations was confirmed by sequencing.
Plasmid pcDNA4/To/Myc-PML II K/SIM mut was then transfected into the HEp-2 TetOn cells (24). Stably transfected colonies were selected in HEp-2 growth medium containing 6 μg/ml of blasticidin and 100 μg/ml of zeocin. Stable cell lines were then identified by Western blotting after a 24-h doxycycline induction.
PML half-life assay.
HEp-2 TetOn and U2OS Tet-On cells stably expressing PML isoforms were seeded in 35-mm plates at ∼50% confluence. Cells were then induced with 1 μg/ml doxycycline for 24 h and infected by recombinant viruses at 10 PFU/cell, as indicated in the Results section. After 1 h of exposure to the virus, inocula were removed and cells were incubated in growth medium supplemented with 10% newborn calf serum for 1 h before adding 100 μg/ml cycloheximide. The time cycloheximide was added was considered 0 hour. Then the infected cells are harvest at 2 h intervals and total cell lysates were then subjected to Western blotting. Band intensity for myc-tagged PML from duplicated experiments was quantitated by ImageJ software and the half-life of PML with standard error was plotted by Microsoft Excel.
In vivo PML ubiquitination assay.
HEp-2 TetOn cells expressing PML II or PML II K/SIM mutant seeded in 100-mm plates were induced with 1 μg/ml doxycycline for 24 h before being exposed to a recombinant virus at 5 PFU/cell. At 3 hpi, cells were scraped, washed, and lysed in urea lysis buffer (8 M urea, 100 mM Na2HPO4/NaH2PO4, pH 8.0, 10 mM Tris-HCl, pH 8.0, 5 mM imidazole, 10 mM β-mercaptoethonal, 50 μM deubiquitinase inhibitor PR619, and 1× protease inhibitor cocktail [Sigma]). Then the cell lysates were subjected to brief sonication before spinning down at 14,000 rpm for 5 min. The supernatant was incubated with Ni-NTA agarose beads (Qiagen) for 4 h at room temperature with gentle agitation. The beads were then washed four times for 5 min each as follows: first with urea lysis buffer; second with urea wash buffer (8 M urea, 100 mM Na2HPO4/NaH2PO4, 10 mM Tris-HCl, pH 6.8, 15 mM imidazole, 10 mM β-mercaptoethonal); third with urea wash buffer + 0.1% Triton X-100; last with PBS. After washing, the HisMyc-PML II was eluted with elution buffer (0.2 M imidazole, 0.15 M Tris-HCl [pH 6.8], 30% glycerol, 0.72 M β-mercaptoethonal, 5% SDS). The eluates were then subjected to Western blotting.
Western blotting.
Protein samples from the half-life assay or the in vivo ubiquitination assay were electrophoretically separated on SDS-PAGE gels and then transferred onto a polyvinylidene difluoride (PVDF) membrane (Millipore). The membrane was blocked with 1× Tris-buffered saline–Tween (TBST) (20 mM Tris [pH 7.5], 150 mM NaCl, 0.5% Tween 20) containing 5% nonfat dry milk and probed with primary antibodies as indicated in the Results section. The membrane was then incubated with horseradish peroxidase-conjugated goat anti-mouse or goat anti-rabbit secondary antibody (Sigma) and visualized with ECL detection reagent (GE Healthcare).
Confocal microscopy.
HEp-2 TetOn cells expressing PML II or PML II K/SIM mutant grown on four-well glass slides (Thermo Fisher Scientific) were uninduced or induced with 1 μg/ml doxycycline for 24 h. Cells were then fixed in 4% paraformaldehyde, permeabilized with 0.2% Triton X-100, and blocked with PBS containing 5% horse serum and 1% bovine serum albumin. Cells were reacted with primary antibodies at 4°C overnight, rinsed and reacted with FITC-conjugated goat anti-rabbit (Sigma) plus Texas Red-conjugated goat anti-mouse (Invitrogen) secondary antibodies. Slides were then mounted with VectaShield (Vector Laboratories) and images were taken with a Leica TCS SP8 confocal microscope.
Antibodies.
An anti-ICP0 polyclonal antibody raised against ICP0 exon 2 has been described elsewhere (46). Anti-PML polyclonal antibody, anti-Myc monoclonal antibody, and anti-actin monoclonal antibody were purchased from Santa Cruz Biotechnology, Inc. Both mono- and polyclonal anti-mCherry antibodies were purchased from Clontech. An anti-ubiquitin polyclonal antibody was purchased from Abcam. Anti-Sp100 and anti-Daxx polyclonal antibodies were purchased from Sigma.
Titration to measure viral growth.
HEp-2 TetOn cells expressing either wild-type PML II or PML II K/SIMmt were seeded in 35-mm plates at ∼50% confluence before being mock-induced or inducted by 1 μg/ml doxycycline for 24 h. Triplicate plates of cells were then infected with the indicated viruses at 0.1 PFU/cell and the inocula were replaced with growth medium after 1 h. Infected cells were harvested at the indicated time points and then lysed and titrated on U2OS cells.
qPCR.
HEp-2 TetOn cells expressing wild-type PML II grown on 35-mm plates were induced with 1 μg/ml doxycycline overnight before being infected with recombinant viruses at 0.1 PFU/cell for 1 h. The inocula were removed, and cells were incubated in growth medium supplemented with 10% newborn calf serum. At 2 and 24 hpi, total DNA was extracted and subjected to quantitative PCR (qPCR) as described elsewhere (45). Briefly, primers targeting the ICP27 gene were used for viral DNA quantitation and QuantumRNA.
A universal 18S internal standard (Invitrogen) was used to normalize total cell DNA. The viral DNA fold increase from 2 hpi to 24 hpi was calculated by comparative threshold cycle (ΔΔCT) analysis.
ACKNOWLEDGMENTS
We are grateful for financial support from NIH grant RO1AI118992 and the ReBUILDetroit pilot project fund awarded to Haidong Gu.
We thank Zackary Kreza for his help in constructing mutations in PML II. We also thank the Microscopy, Imaging and Cytometry Resources (MICR) Core facility at Wayne State University for technical support.
REFERENCES
- 1.Roizman B, Knipe DM, Whitley RJ. 2013. Herpes simplex viruses, p 1823–1897. In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B (ed), Fields virology, 6th ed, vol 2Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 2.Gu H. 2016. Infected cell protein 0 functional domains and their coordination in herpes simplex virus replication. World J Virol 5:1–13. doi: 10.5501/wjv.v5.i1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chelbi-Alix MK, de Thé H. 1999. Herpes virus induced proteasome-dependent degradation of the nuclear bodies-associated PML and Sp100 proteins. Oncogene 18:935–941. doi: 10.1038/sj.onc.1202366. [DOI] [PubMed] [Google Scholar]
- 4.Parkinson J, Lees-Miller SP, Everett RD. 1999. Herpes simplex virus type 1 immediate-early protein vmw110 induces the proteasome-dependent degradation of the catalytic subunit of DNA-dependent protein kinase. J Virol 73:650–657. doi: 10.1128/JVI.73.1.650-657.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Orzalli MH, DeLuca NA, Knipe DM. 2012. Nuclear IFI16 induction of IRF-3 signaling during herpesviral infection and degradation of IFI16 by the viral ICP0 protein. Proc Natl Acad Sci U S A 109:E3008–3017. doi: 10.1073/pnas.1211302109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chaurushiya MS, Lilley CE, Aslanian A, Meisenhelder J, Scott DC, Landry S, Ticau S, Boutell C, Yates JR 3rd, Schulman BA, Hunter T, Weitzman MD. 2012. Viral E3 ubiquitin ligase-mediated degradation of a cellular E3: viral mimicry of a cellular phosphorylation mark targets the RNF8 FHA domain. Mol Cell 46:79–90. doi: 10.1016/j.molcel.2012.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Everett RD, Parada C, Gripon P, Sirma H, Orr A. 2008. Replication of ICP0-null mutant herpes simplex virus type 1 is restricted by both PML and Sp100. J Virol 82:2661–2672. doi: 10.1128/JVI.02308-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Merkl PE, Orzalli MH, Knipe DM. 2018. Mechanisms of host IFI16, PML, and Daxx protein restriction of herpes simplex virus 1 replication. J Virol 92. doi: 10.1128/JVI.00057-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lallemand-Breitenbach V, de Thé H. 2018. PML nuclear bodies: from architecture to function. Curr Opin Cell Biol 52:154–161. doi: 10.1016/j.ceb.2018.03.011. [DOI] [PubMed] [Google Scholar]
- 10.Chang HR, Munkhjargal A, Kim MJ, Park SY, Jung E, Ryu JH, Yang Y, Lim JS, Kim Y. 2018. The functional roles of PML nuclear bodies in genome maintenance. Mutat Res 809:99–107. doi: 10.1016/j.mrfmmm.2017.05.002. [DOI] [PubMed] [Google Scholar]
- 11.Gu H, Zheng Y. 2016. Role of ND10 nuclear bodies in the chromatin repression of HSV-1. Virol J 13:62. doi: 10.1186/s12985-016-0516-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fagioli M, Alcalay M, Pandolfi PP, Venturini L, Mencarelli A, Simeone A, Acampora D, Grignani F, Pelicci PG. 1992. Alternative splicing of PML transcripts predicts coexpression of several carboxy-terminally different protein isoforms. Oncogene 7:1083–1091. [PubMed] [Google Scholar]
- 13.Jensen K, Shiels C, Freemont PS. 2001. PML protein isoforms and the RBCC/TRIM motif. Oncogene 20:7223–7233. doi: 10.1038/sj.onc.1204765. [DOI] [PubMed] [Google Scholar]
- 14.Wang P, Benhenda S, Wu H, Lallemand-Breitenbach V, Zhen T, Jollivet F, Peres L, Li Y, Chen S-J, Chen Z, de Thé H, Meng G. 2018. RING tetramerization is required for nuclear body biogenesis and PML sumoylation. Nat Commun 9:1277. doi: 10.1038/s41467-018-03498-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Banani SF, Rice AM, Peeples WB, Lin Y, Jain S, Parker R, Rosen MK. 2016. Compositional control of phase-separated cellular bodies. Cell 166:651–663. doi: 10.1016/j.cell.2016.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Van Damme E, Laukens K, Dang TH, Van Ostade X. 2010. A manually curated network of the PML nuclear body interactome reveals an important role for PML-NBs in SUMOylation dynamics. Int J Biol Sci 6:51–67. doi: 10.7150/ijbs.6.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Komatsu T, Nagata K, Wodrich H. 2016. The role of nuclear antiviral factors against invading DNA viruses: the immediate fate of incoming viral genomes. Viruses 8:290. doi: 10.3390/v8100290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gu H, Roizman B. 2007. Herpes simplex virus-infected cell protein 0 blocks the silencing of viral DNA by dissociating histone deacetylases from the CoREST-REST complex. Proc Natl Acad Sci U S A 104:17134–17139. doi: 10.1073/pnas.0707266104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ferenczy MW, Ranayhossaini DJ, Deluca NA. 2011. Activities of ICP0 involved in the reversal of silencing of quiescent herpes simplex virus 1. J Virol 85:4993–5002. doi: 10.1128/JVI.02265-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gu H, Roizman B. 2009. The two functions of herpes simplex virus 1 ICP0, inhibition of silencing by the CoREST/REST/HDAC complex and degradation of PML, are executed in tandem. J Virol 83:181–187. doi: 10.1128/JVI.01940-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boutell C, Cuchet-Lourenco D, Vanni E, Orr A, Glass M, McFarlane S, Everett RD. 2011. A viral ubiquitin ligase has substrate preferential SUMO targeted ubiquitin ligase activity that counteracts intrinsic antiviral defence. PLoS Pathog 7:e1002245. doi: 10.1371/journal.ppat.1002245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao X. 2018. SUMO-mediated regulation of nuclear functions and signaling processes. Mol Cell 71:409–418. doi: 10.1016/j.molcel.2018.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ribet D, Cossart P. 2018. Ubiquitin, SUMO, and NEDD8: key targets of bacterial pathogens. Trends Cell Biol 28:926–940. doi: 10.1016/j.tcb.2018.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zheng Y, Samrat SK, Gu H. 2016. A tale of two PMLs: elements regulating a differential substrate recognition by the ICP0 E3 ubiquitin ligase of herpes simplex virus 1. J Virol 90:10875–10885. doi: 10.1128/JVI.01636-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kwon YT, Ciechanover A. 2017. The ubiquitin code in the ubiquitin-proteasome system and autophagy. Trends Biochem Sci 42:873–886. doi: 10.1016/j.tibs.2017.09.002. [DOI] [PubMed] [Google Scholar]
- 26.Rape M. 2018. Ubiquitylation at the crossroads of development and disease. Nat Rev Mol Cell Biol 19:59–70. doi: 10.1038/nrm.2017.83. [DOI] [PubMed] [Google Scholar]
- 27.Ishov AM, Maul GG. 1996. The periphery of nuclear domain 10 (ND10) as site of DNA virus deposition. J Cell Biol 134:815–826. doi: 10.1083/jcb.134.4.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gu H, Roizman B. 2009. Engagement of the lysine-specific demethylase/HDAC1/CoREST/REST complex by herpes simplex virus 1. J Virol 83:4376–4385. doi: 10.1128/JVI.02515-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tatham MH, Geoffroy MC, Shen L, Plechanovova A, Hattersley N, Jaffray EG, Palvimo JJ, Hay RT. 2008. RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nat Cell Biol 10:538–546. doi: 10.1038/ncb1716. [DOI] [PubMed] [Google Scholar]
- 30.Everett RD, Meredith M, Orr A, Cross A, Kathoria M, Parkinson J. 1997. A novel ubiquitin-specific protease is dynamically associated with the PML nuclear domain and binds to a herpesvirus regulatory protein. EMBO J 16:1519–1530. doi: 10.1093/emboj/16.7.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boutell C, Everett RD. 2013. Regulation of alphaherpesvirus infections by the ICP0 family of proteins. J Gen Virol 94:465–481. doi: 10.1099/vir.0.048900-0. [DOI] [PubMed] [Google Scholar]
- 32.Newhart A, Rafalska-Metcalf IU, Yang T, Negorev DG, Janicki SM. 2012. Single cell analysis of Daxx and ATRX-dependent transcriptional repression. J Cell Sci 125:5489–5501. doi:10.1242/jcs.110148. doi: 10.1242/jcs.110148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deschamps T, Kalamvoki M. 2017. Impaired STING pathway in human osteosarcoma U2OS cells contributes to the growth of ICP0-null mutant herpes simplex virus. J Virol 91:e00006-17. doi: 10.1128/JVI.00006-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yao F, Schaffer PA. 1995. An activity specified by the osteosarcoma line U2OS can substitute functionally for ICP0, a major regulatory protein of herpes simplex virus type 1. J Virol 69:6249–6258. doi: 10.1128/JVI.69.10.6249-6258.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Everett RD, Meredith M, Orr A. 1999. The ability of herpes simplex virus type 1 immediate-early protein Vmw110 to bind to a ubiquitin-specific protease contributes to its roles in the activation of gene expression and stimulation of virus replication. J Virol 73:417–426. doi: 10.1128/JVI.73.1.417-426.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boutell C, Canning M, Orr A, Everett RD. 2005. Reciprocal activities between herpes simplex virus type 1 regulatory protein ICP0, a ubiquitin E3 ligase, and ubiquitin-specific protease USP7. J Virol 79:12342–12354. doi: 10.1128/JVI.79.19.12342-12354.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kalamvoki M, Gu H, Roizman B. 2012. Overexpression of the ubiquitin-specific protease 7 resulting from transfection or mutations in the ICP0 binding site accelerates rather than depresses herpes simplex virus 1 gene expression. J Virol 86:12871–12878. doi: 10.1128/JVI.01981-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seeler JS, Dejean A. 2001. SUMO: of branched proteins and nuclear bodies. Oncogene 20:7243–7249. doi: 10.1038/sj.onc.1204758. [DOI] [PubMed] [Google Scholar]
- 39.Shen TH, Lin HK, Scaglioni PP, Yung TM, Pandolfi PP. 2006. The mechanisms of PML-nuclear body formation. Mol Cell 24:331–339. doi: 10.1016/j.molcel.2006.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pichler A, Fatouros C, Lee H, Eisenhardt N. 2017. SUMO conjugation—a mechanistic view. Biomol Concepts 8:13–36. doi: 10.1515/bmc-2016-0030. [DOI] [PubMed] [Google Scholar]
- 41.Yu H, Chen JK, Feng S, Dalgarno DC, Brauer AW, Schreiber SL. 1994. Structural basis for the binding of proline-rich peptides to SH3 domains. Cell 76:933–945. doi: 10.1016/0092-8674(94)90367-0. [DOI] [PubMed] [Google Scholar]
- 42.Lim WA, Richards FM, Fox RO. 1994. Structural determinants of peptide-binding orientation and of sequence specificity in SH3 domains. Nature 372:375–379. doi: 10.1038/372375a0. [DOI] [PubMed] [Google Scholar]
- 43.Banani SF, Lee HO, Hyman AA, Rosen MK. 2017. Biomolecular condensates: organizers of cellular biochemistry. Nat Rev Mol Cell Biol 18:285–298. doi: 10.1038/nrm.2017.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zheng Y, Gu H. 2015. Identification of three redundant segments responsible for herpes simplex virus 1 ICP0 to fuse with ND10 nuclear bodies. J Virol 89:4214–4226. doi: 10.1128/JVI.03658-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gu H, Zheng Y, Roizman B. 2013. The interaction of herpes simplex virus ICP0 with ND10 bodies: a sequential process of adhesion, fusion and retention. J Virol 87:10244–10254. doi: 10.1128/JVI.01487-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Samrat SK, Ha BL, Zheng Y, Gu H. 2017. Characterization of elements regulating the nuclear-to-cytoplasmic translocation of ICP0 in late herpes simplex virus 1 infection. J Virol 9287:e01673-17. doi: 10.1128/JVI.01673-17. [DOI] [PMC free article] [PubMed] [Google Scholar]