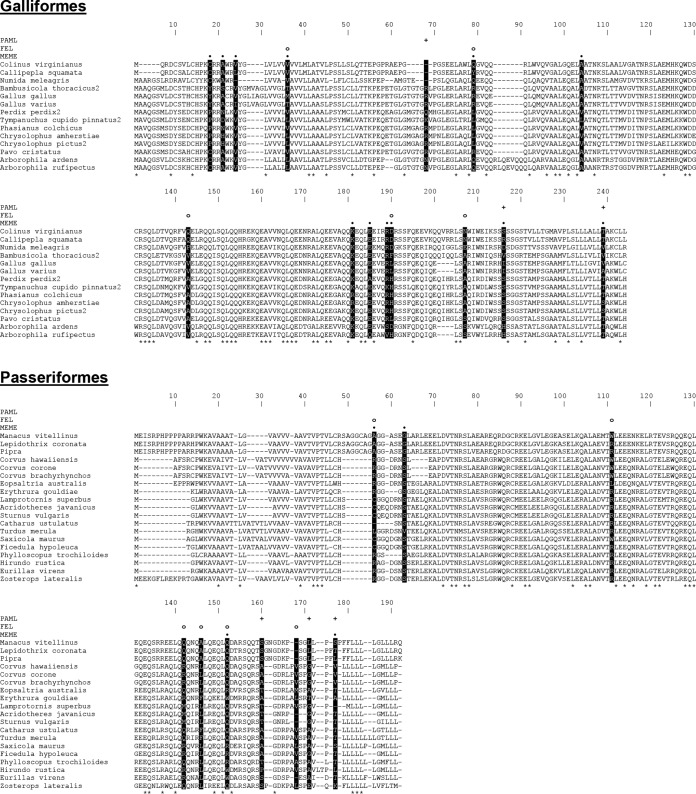
FIG 7.
Positively selected sites detected in avian BST-2 orthologs by three different algorithms. Amino acid alignments of Galliformes (top) and Passeriformes (bottom) BST-2 are shown. Residues with 100% conservation in each alignment are marked with asterisks below the sequences. Positively selected sites are highlighted as black columns. Predictions of the individual algorithms are depicted above the alignment as plus signs (PAML), open circles (FEL), and black dots (MEME).