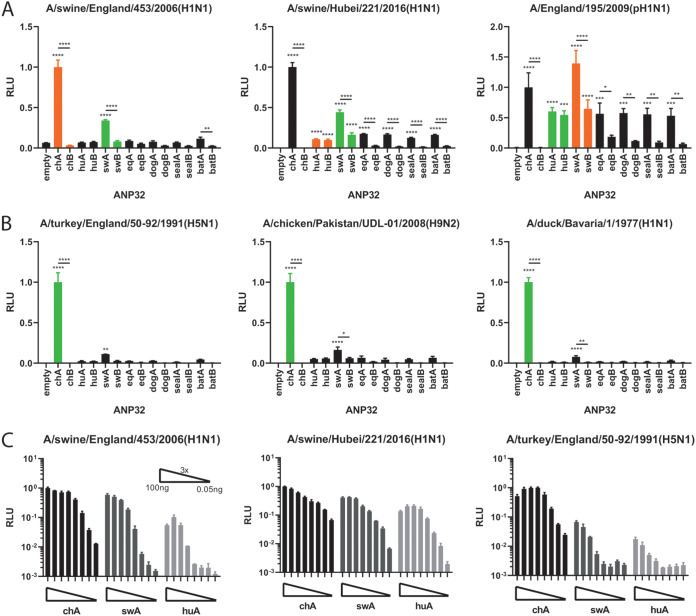
FIG 2.
swANP32A can support the activity of minimally mammalian-adapted or completely nonadapted polymerases. Minigenome assays of swine (A) and avian (B) polymerases performed in human eHAP dKO cells with ANP32 proteins from different avian or mammalian species cotransfected. Green bars indicate species from which the influenza virus polymerase was isolated; orange bars indicate recent species from which the virus has jumped. Data indicate triplicate repeats plotted as mean with standard deviation. Data for each polymerase normalized to chicken ANP32A. (C) ANP32 protein titrations with three different virus polymerase constellations. ANP32 expression plasmids were diluted in a series of 3× dilutions starting with 100 ng. Data indicate triplicate repeats plotted as mean with standard deviation. Statistical significance was determined by one-way analysis of variance (ANOVA) with multiple comparisons against empty vector. **, 0.01 ≥ P > 0.001; ***, 0.001 ≥ P > 0.0001; ****, P ≤ 0.0001.