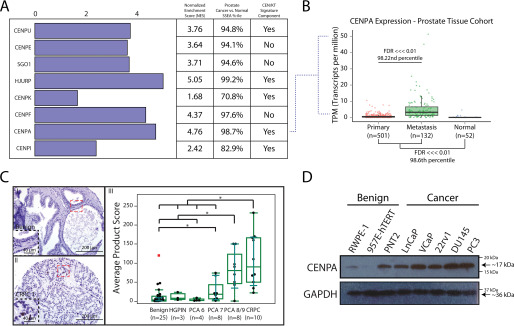
Figure 1.
Overexpression of CENPA in prostate cancer. A, SSEA was used to query a catalogue of curated RNA-seq libraries (n = 685) for differentially expressed centromeric genes in the prostate tissue type cohort. Genes were selected based on associations identified in prior studies with cancer progression and were characterized by their inclusion in the previously described CEN/KT signature that negatively impacts therapy response and survival. B, focused SSEA on CENPA mRNA levels depicted as transcripts per million (TPM) in normal prostate (n = 52), primary prostate cancer (n = 501), and metastatic prostate cancer (n = 132) tissue. C, tissue microarray (n = 58 total tissues, n = 174 cores) of benign prostate (I), high-grade prostatic intraepithelial neoplasia (HGPIN), Gleason grade 6–9 prostate cancer (PCA), and castration-resistant prostate cancer (CRPC) (II) tissue stained for CENPA. *, P < 0.05. Staining was evaluated by assessing the most frequent pattern of intensity at 20× and the percentage of cells exhibiting that pattern (III). D, immunoblot for CENPA and GAPDH (loading control) in a panel of benign and malignant prostate cell lines. Note that PNT2, although benign, proliferates the most rapidly of all cell lines tested (see also Fig. S1C).