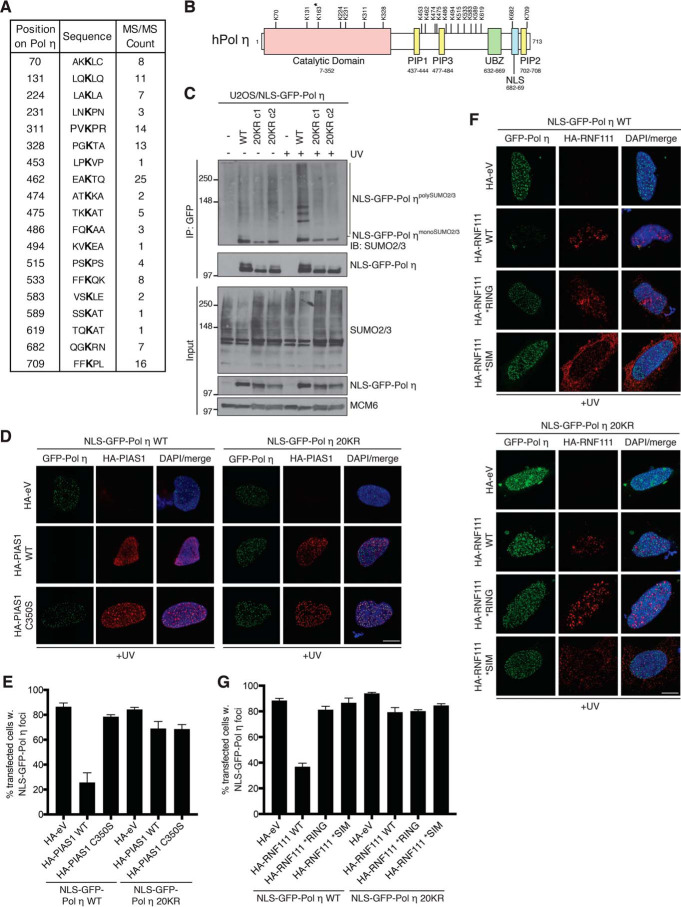
Figure 4.
Multisite SUMOylation of Pol η underlies its PIAS1- and RNF111-dependent extraction from DNA damage sites. A, table showing identification of endogenous Pol η SUMOylation sites using an augmented K0-SUMO proteomic strategy coupled with deep MS data analysis. See also Table S3 and Fig. S4. B, schematic diagram of human Pol η, depicting the location of functional motifs and identified SUMOylation sites. A previously reported Pol η SUMOylation site, Lys-163 (37), which did not pass the significance threshold but was mutated along with the 19 high-confidence SUMOylation sites to generate the Pol η 20KR mutant, is indicated by the asterisk. C, U2OS cells or derivative clones stably expressing GFP-Pol η WT or 20KR mutant (clone 1 (c1) and clone 2 (c2)) were left untreated or exposed to UV, lysed, and subjected to GFP IP under denaturing conditions and immunoblotted (IB) with the indicated antibodies. D, representative images of U2OS cells co-transfected with the indicated GFP-Pol η and HA-PIAS1 expression constructs, exposed to UV and immunostained with HA antibody. Scale bar, 10 μm. eV, empty vector. E, quantification of data in D (mean ± S.E.M.; n = 3 independent experiments; ≥55 cells analyzed per condition). F, representative images of U2OS cells co-transfected with the indicated GFP-Pol η and HA-RNF111 expression constructs, exposed to UV, and immunostained with HA antibody. Scale bar, 10 μm. G, quantification of data in F (mean ± S.E.M.; n = 3 independent experiments; ≥100 cells analyzed per condition).