Figure 1.
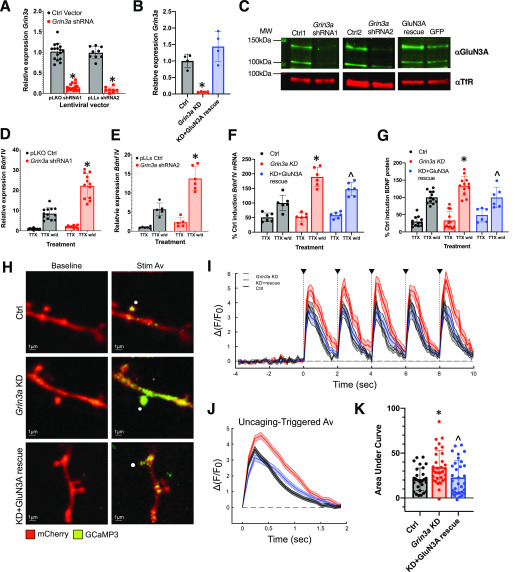
GluN3A knockdown selectively enhances NMDAR-dependent Bdnf transcription. A, Levels of Grin3a mRNA in rat hippocampal neurons infected with lentiviruses encoding shRNAs targeting Grin3a (shRNA1 and shRNA2) or their paired control vectors (pLKO or pLLx3.8). Grin3a mRNA levels are scaled to levels in cells infected with the paired control viruses. Two-way ANOVA for vector, F(1, 45) = 1.033, p = 0.32; and shRNA, F(1, 45) = 425.6, p < 0.0001. For pLKO shRNA1 versus Ctrl1, n = 16/virus, p < 0.0001. For pLLx3.8 shRNA2 versus Ctrl2, n = 9 Ctrl2, 8 shRNA2, p < 0.0001. B, Expression of Grin3a mRNA in neurons infected with the Grin3a KD and shRNA-resistant GluN3A rescue viruses. C, Membrane fraction from cultured hippocampal neurons infected with the indicated lentiviruses, analyzed by Western blotting using an antibody that detects the GluN3A protein (green). Transferrin receptor is shown as a loading control (red). The predicted molecular weight (MW) of GluN3A is 130 kDa. D and E, Levels of Bdnf IV mRNA in hippocampal neurons infected with the indicated lentiviruses in the presence of TTX or 6 h following TTX w/d. Values are scaled to Ctrl TTX for comparison. D, pLKO. Two-way ANOVA for treatment, F(1, 41) = 49.53, p < 0.0001; for virus, F(1, 41) = 172.5, p < 0.0001; and for treatment–virus interaction, F(1, 41) = 36.12, p < 0.0001. Ctrl TTX, n = 11; Ctrl TTX w/d, n = 13; shRNA TTX, n = 9; shRNA TTX w/d, n = 12. Ctrl TTX versus TTX w/d, p < 0.0001; shRNA TTX versus TTX w/d, p < 0.0001. Ctrl TTX w/d versus shRNA TTX w/d, p < 0.0001. Ctrl TTX versus shRNA TTX is not significant, p = 0.90. E, pLLx3.8. Two-way ANOVA for treatment, F(1, 18) = 32.97, p < 0.0001; for virus, F(1, 18) = 97.67, p < 0.0001; and for treatment–virus interaction, F(1, 18) = 16.98, p = 0.0006. Ctrl TTX, n = 6; Ctrl TTX w/d, n = 6; shRNA TTX, n = 6; shRNA TTX w/d, n = 5. Ctrl TTX versus TTX w/d, p = 0.0036; shRNA TTX versus TTX w/d, p < 0.0001. Ctrl TTX w/d versus shRNA TTX w/d, p < 0.0001. Ctrl TTX versus shRNA TTX is not significant, p = 0.67. F, Levels of TTX w/d-induced Bdnf IV mRNA in hippocampal neurons infected with the indicated lentiviruses relative to the control condition (Ctrl), whose induced average is scaled to 100%. Two-way ANOVA for treatment, F(1, 30) = 160.5, p < 0.0001; for virus, F(2, 30) = 13.33, p = 0.0001; and for treatment–virus interaction, F(2, 30) = 12.25, p = 0.0001. n = 6/condition. For TTX w/d, Ctrl versus KD, p < 0.0001; KD versus rescue, p = 0.0064; Ctrl versus rescue, p = 0.0019. G, Levels of BDNF protein in hippocampal neurons infected with the indicated lentiviruses in the presence of TTX or 6 h following TTX w/d. Control condition (Ctrl) TTX w/d induced average is scaled to 100%. Two-way ANOVA for treatment, F(1, 54) = 144.8, p < 0.0001; for virus, F(2, 54) = 4.103, p = 0.022; and for treatment–virus interaction, F(2, 54) = 5.547, p = 0.0064. n = 12/condition for Ctrl and KD, 6/condition for rescue. For TTX w/d Ctrl versus Grin3a KD, p = 0.0016; KD versus rescue, p = 0.012. H, Representative 2-photon images of hippocampal CA1 neurons expressing GCaMP3 (green) and mCherry (red). (Left) Baseline images corresponding to the average projection for the first 31 frames of imaging, prior to uncaging. (Right) Average projection images of the first two frames after each of the five uncaging pulses (10 total imaging frames). White circles indicate the uncaging point. (I) Average ΔF/F0 traces showing response to glutamate uncaging (black triangles) for control (n = 29 spines; 3 cells), Grin3a KD (n = 32 spines, 3 cells), and KD plus GluN3A rescue (n = 32 spines, 3 cells) conditions. Data correspond to mean ± S.E. J, Uncaging-triggered average of the responses shown in panel I. Traces correspond to the average baseline-subtracted response to each glutamate uncaging pulse, where the baseline is the frame immediately before each uncaging stimulus. Error corresponds to S.E. of the five uncaging pulses for each condition. K, Average integrated GCaMP3 signal for each condition. The average area under the curve was calculated for each spine and then averaged for a given condition. Asterisks indicate statistical significance as determined by ANOVA and a post hoc test using Fisher's least significant difference. Ctrl versus KD, p = 0.003; ctrl versus rescue, p = 0.6; KD versus rescue, p = 0.009. *, p < 0.05 for Grin3a KD versus Ctrl; ∧, p < 0.05 GluN3A rescue versus Grin3a KD.