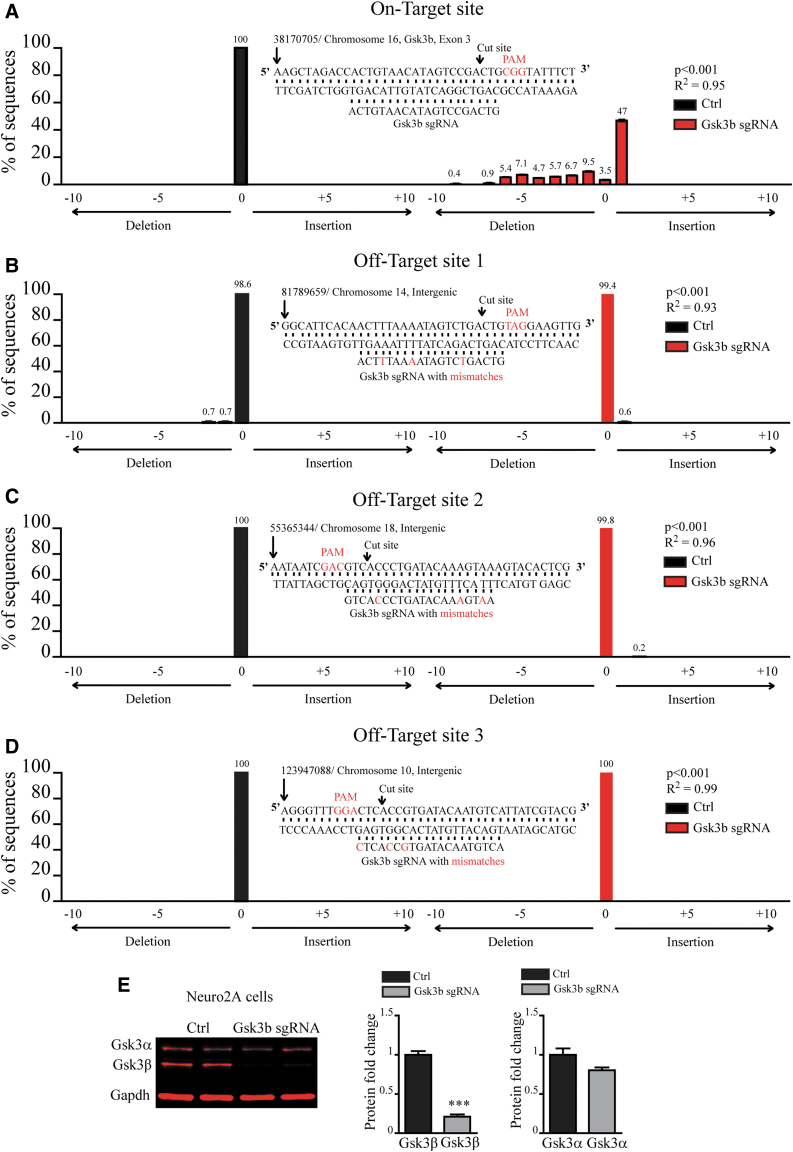
FIG. 1.
Specificity of CRISPR-Cas9-mediated Gsk3β knockout. Quantification of mutations using TIDE analysis at (A) on-target and (B–D) three putative off-target sites. Position “0” represents the percentage of non-mutated sequence, “–” positions are deletions, and “+” positions are insertions. Middle panels show the nucleotide sequence and chromosomal position at a putative target site, Gsk3b sgRNA, as well as mismatch nucleotides at off-target sites. (E) Quantification of the expression of Gsk3α and Gsk3β after CRISPR-Cas9-mediated knockout of Gsk3β in Neuro2A cells (Gsk3α: Ctrl 1 ± 0.08 n = 5, Gsk3b sgRNA 0.8 ± 0.035 n = 4; Gsk3β: Ctrl 1 ± 0.047 n = 5, Gsk3b sgRNA 0.21 ± 0.028 n = 5; *p < 0.05, Student's t-test). Left panel: an example of Western blot membrane stained for Gsk3α, Gsk3β, and Gapdh.