Figure 2.
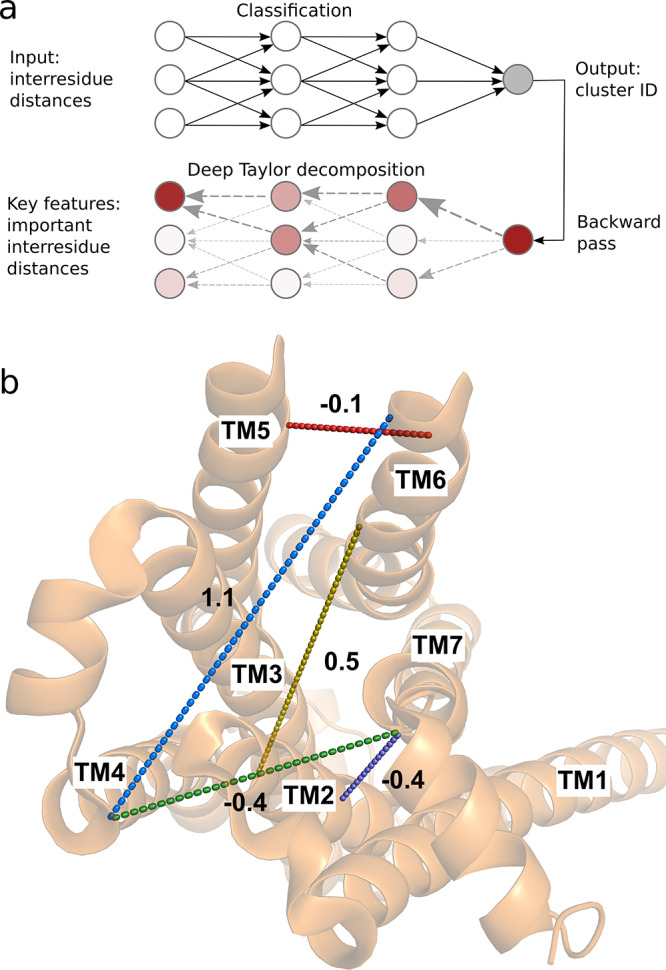
Five-dimensional collective variable (CV) space used to optimize the minimum free energy path identified in a data-driven manner. (a) A fully connected neural network was trained to classify configurations in active, intermediate, and inactive metastable states (clusters). Deep Taylor decomposition was then used to identify the most important input inter-residue distances for the classification decision. The top-ranked distances were used as CVs. (b) The five CVs used in this work projected onto an intracellular view of the active crystal structure (PDB entry 3P0G). The CVs corresponding to TM2–TM7, TM6–TM4, TM7–TM4, TM3–TM6, and TM6–TM5 distances defined in Table S2 are shown as purple, blue, green, yellow, and red dashed lines, respectively. The change of these distance CVs from the inactive to the active state structures is reported in nanometers.
