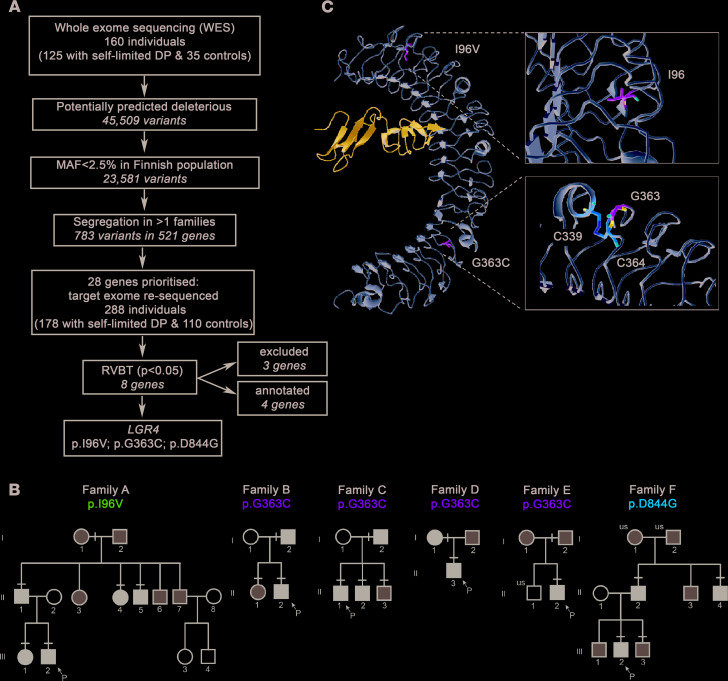
Figure 1. Identification of LGR4 as a candidate gene for self-limited DP with rare pathogenic variants in patients.
(A) Whole-exome sequencing (WES) was performed on 160 individuals from our cohort (125 with self-limited DP and 35 controls). Variants were filtered using filters for quality control, predicted functional annotation, minor allele frequency (MAF), and for genes with variants in multiple families. A total of 28 genes were prioritized and were targeted exome sequenced in additional 288 individuals. Further analysis identified genes significantly enriched for pathogenic variants via whole gene burden testing, and genes involved in GnRH neuronal development and puberty timing (1, 2, 10, 11). Excluded, owing to the presence of variants in multiple controls. (B) Squares and circles indicate male and female family members, respectively. Black symbols represent affected individuals, gray symbols represent unknown phenotype, and clear symbols represent unaffected individuals. “P” indicates the proband in each family, and “us” indicates unsequenced owing to lack of DNA. A black line above an individual’s symbol indicates heterozygosity for that mutation as confirmed by either WES or Fluidigm array, and verified by Sanger sequencing. (C) LGR4 extracellular domain (gold) with variants bound to R-spondin1 (blue). Variants p.I96V and p.G363C are presented (green). p.I96V and p.G363C are in the variable region of LRR2 and LRR12, respectively. p.G363C occurs in close proximity to a cysteine bond (C339-C364; orange), and this substitution introduces a steric clash. p.D844G is within the cytoplasmic domain, and no experimental structure for the LGR4 cytoplasmic domain was available. DP, delayed puberty.