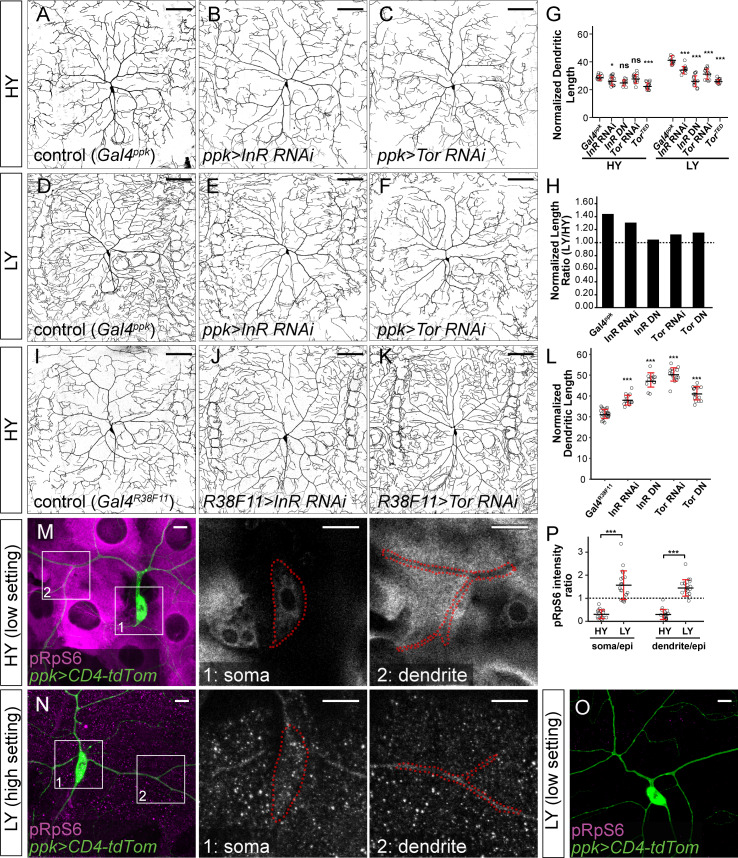
Figure 2. The InR-Tor pathway underlies the preferential neuronal growth under nutrient stress.
(A–F) ddaC neurons in the Gal4ppk control (A and D) and animals expressing Gal4ppk-driven InR RNAi (B and E) and Tor RNAi (C and F) in HY and LY conditions. (G) Quantification of normalized dendrite length (total dendrite length/segment width) in HY and LY conditions. HY: n = 14 for Gal4ppk, n = 15 for InR RNAi, n = 11 for InR DN, n = 15 for Tor RNAi, n = 15 for Tor DN; LY: n = 14 for Gal4ppk, n = 12 for InR RNAi, n = 14 for InR DN, n = 15 for Tor RNAi, n = 15 for Tor DN. Two-way ANOVA, Posthoc contrasts with a Dunnett correction. The differences between control and InR RNAi under HY and LY conditions are significantly different as indicated by a significant interaction term (p=0.003924), the same for InR DN (p=8.581e-09), Tor RNAi (p=5.876e-07) and Tor DN (p=1.062e-09). (H) The ratios of average normalized dendrite length between LY and HY. (I–K) ddaC neurons in the Gal4R38F11 control (I) and animals expressing Gal4R38F11-driven InR RNAi (J) and Tor RNAi (K) in HY condition. (L) Quantification of normalized dendrite length in e Gal4R38F11-driven knockdown and overexpression. n = 23 for Gal4R38F11, n = 17 for InR RNAi, n = 17 for InR DN, n = 16 for Tor RNAi, n = 17 for Tor DN. One-way ANOVA and Tukey’s HSD test. (M–O) pRpS6 staining (magenta) of ddaC neurons (Green) and epidermal cells in HY and LY conditions in 2-dimensional (2D) projections. The insets in (M) and (N) show pRpS6 staining at the soma (1) and primary dendrites (2) in single confocal sections, with the somas and dendrites outlined. High settings and low settings stand for high and low pRpS6 detection settings. (P) Quantification of pRpS6 intensity ratio (soma/epidermal cells and dendrites/epidermal cells) in HY and LY conditions. Soma/epi: n = 17 for HY, n = 20 for LY; dendrites/epi: n = 16 for HY, n = 20 for LY. Two-way ANOVA. The differences between HY and LY in soma and dendrites are not significantly different as indicated by a non-significant interaction term (p=0.5592). For all quantifications, ***p<0.001; *p<0.05; ns, not significant. each circle represents a neuron. Significance level is for comparison between the control and the genotype indicated under the same food condition. Black bars, mean; red bars, SD. Scale bars, 100 μm in (A–K); 10 μm in (M–O).