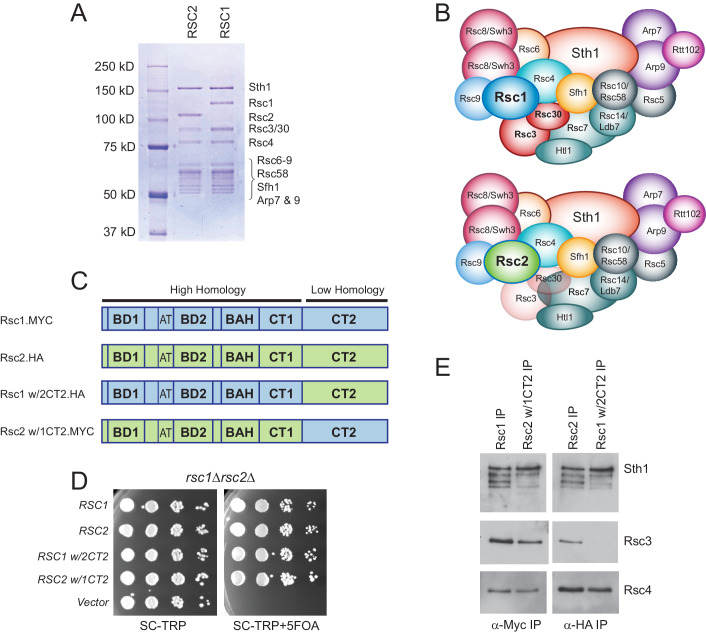
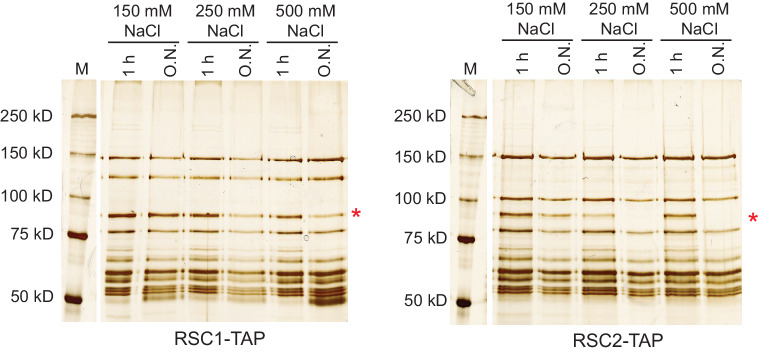
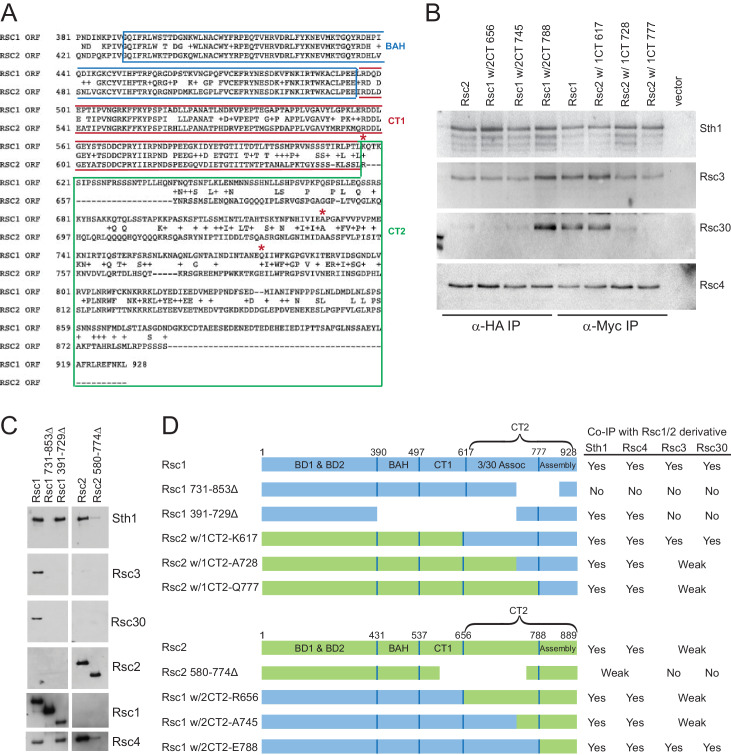
Figure 1. Rsc3/30 module has higher avidity for the RSC1 complex.
(A) Purified RSC1-TAP and RSC2-TAP complexes (2 µg) analyzed on 7.5% SDS-PAGE gel stained with Coomassie dye. (B) RSC1 and RSC2 complex compositions, with decreased opacity displaying the reduced association of the Rsc3/30 module in the RSC2 complex. (C) Domain structure and swaps of Rsc1 and Rsc2. (D) CT2 domain swaps complement for viability. TRP1-marked plasmids bearing RSC1 (p609), RSC2 (p604), RSC1 w/2CT2 (p3097), RSC2 w/1CT2 (p3098), or vector (pRS314) were transformed into rsc1∆ rsc2∆ [RSC1.URA3] (YBC800), and spotted as 10x serial dilutions to SC-TRP or to SC-TRP+5FOA to force the loss of the RSC1.URA3 plasmid. ’ w/’ indicates ‘with’. One of four biological replicates shown. (E) The Rsc3/30 module associates more strongly with the CT2 region of Rsc1. Immunoprecipitations of Rsc1, Rsc2 w/1CT2, Rsc2, Rsc1 w/2CT2 from whole cell extracts. Blots were probed with anti-Sth1, then stripped and reprobed for anti-Rsc3 and anti-Rsc4. One of three technical replicates shown. Figure 1—figure supplement 1. The Rsc3/30 module associates with the RSC1 complex at high stringency. Figure 1—figure supplement 2. Additional swaps and truncations define the region of Rsc3/30 association.