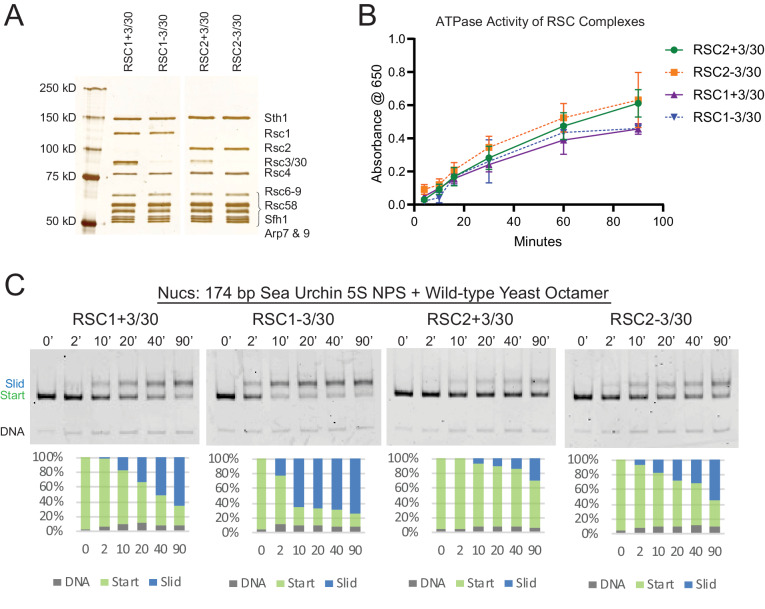
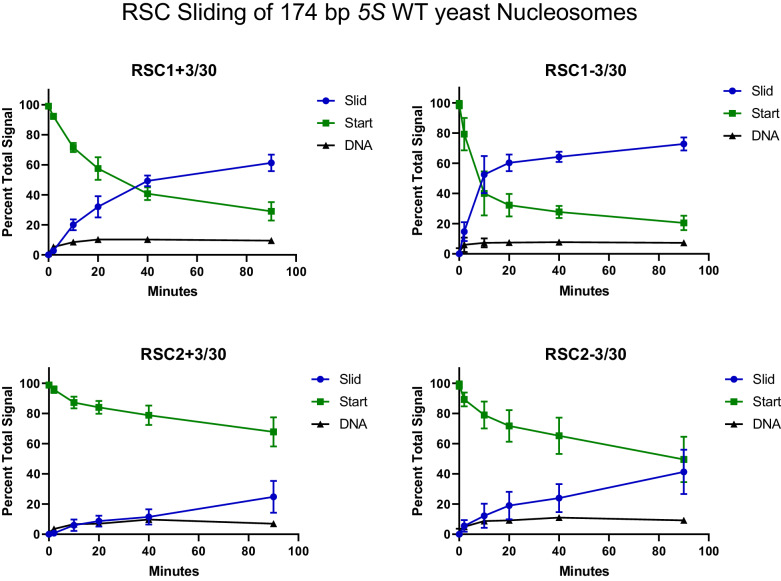
Figure 2. RSC1 and RSC2 complexes differ in remodeling activity on a sea urchin 5S mononucleosomal substrate.
(A) Alternative RSC1 and RSC2 complexes, with the Rsc3/30 module maintained or removed during purification. Purified RSC complexes (600 ng) were analyzed on a 6% polyacrylamide SDS-PAGE gel stained with silver. The RSC1 and RSC2 complexes are from the same gel, but were moved adjacent for the depiction. (B) ATPase time course of RSC1 and RSC2 with and without the Rsc3/30 module. Values are the mean +/- standard deviation from two separate RSC preps for each RSC complex assayed in triplicate. (C) Comparative sliding of 174 bp sea urchin 5S yeast mononucleosomes (20 nM) by RSC1 and RSC2 complexes (30 nM). The nucleosomal Start (green), Slid (blue), and free DNA (grey) bands were quantified and reported as a percent of the total signal.