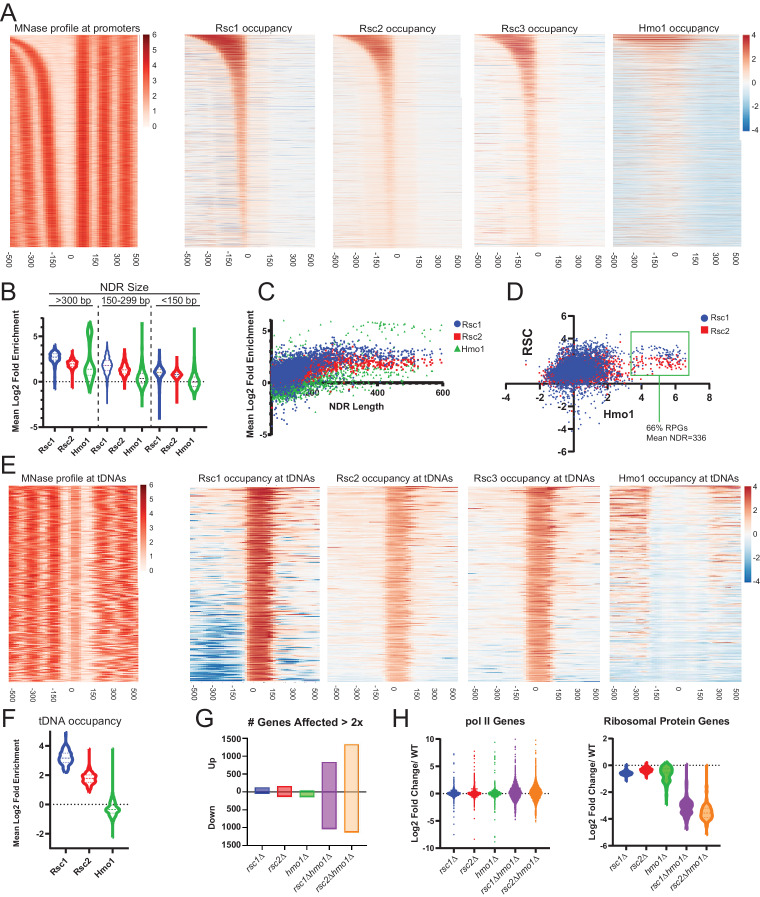
Figure 5. Rsc1 and Rsc2 occupy promoters with wide NDRS, with preferential occupancy of RSC1 at tDNAs.
(A) Heat maps showing enrichment of nucleosomes, Rsc1, Rsc2, Rsc3, and Hmo1 at promoters, sorted by NDR length. (B) Violin plot of Rsc1, Rsc2, and Hmo1 occupancy at promoters at three categories of NDR length. (C) Plot of Rsc1, Rsc2, and Hmo1 occupancy compared to NDR length. (D) Plot of Rsc1 and Rsc2 enrichment by Hmo1 occupancy. (E) Heat maps showing enrichment of nucleosomes, Rsc1, Rsc2, Rsc3, and Hmo1 at all tDNAs (tRNA encoding genes). (F) Violin plot of Rsc1, Rsc2, and Hmo1 mean log2 fold enrichment at tDNAs. (G) Number of genes affected by rsc and hmo1 deletions. For each mutation the number of genes up or downregulated two fold or more compared to WT, rsc1Δ (↑129↓45), rsc2Δ (↑160↓129), hmo1Δ (↑44↓132), rsc1∆ hmo1∆ (↑838↓1028), rsc2∆ hmo1∆ (↑1336↓1131). (H) Gene Expression changes. Violin plots of RNA expression for each mutation at all pol II genes (6145 genes) and at ribosomal protein genes (132 genes) as compared to WT expression. ChIP-seq and RNA-seq data shown represents averages of two and three biological replicates, respectively.