Figure 1.
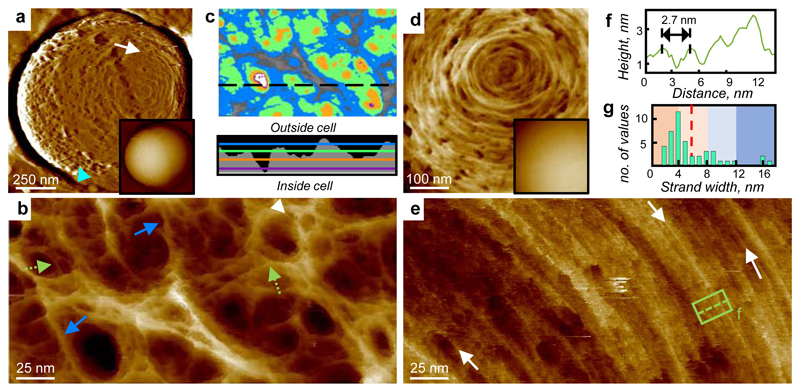
AFM images of peptidoglycan in living S. aureus. a, Entire cell, high-pass filtered (filter size = 0.25 μm, horizontal), blue arrow head: mesh, white arrow: rings. Inset (I): Unfiltered image of the same area. Topographic height data scale (DS) = 902 nm. b, Image of the external surface of mature S. aureus cell wall showing a porous gel structure, white arrow head indicates a feature consistent with a single glycan chain, green dotted arrows examples of strand convergence, and blue arrows multi-strand fibres. DS = 29 nm. c, Depth analysis of pores including the area of image b (colour scale: Grey 0 nm, Blue = 5 nm, Green = 10 nm, Orange = 15 nm, Purple = 20 nm, White = 23 nm). Profile shows a section through the image at the black dotted line on c, coloured lines indicate depths as above, black area shows outline of pores. d, Ring architecture. DS = 22 nm. Inset (I): raw data. DS = 476 nm. e, Dense rings, white arrows indicating examples of likely individual chains. DS = 23 nm. f, Cross-section showing spacing between strands from boxed area in e. g, Histogram of strand width, full width at half maximum (FWHM) from image b, n = 60 strands, dotted line shows mean value. Images b, d and e have been plane-fitted to remove cell curvature. For sample size and data reproducibility, see Methods.