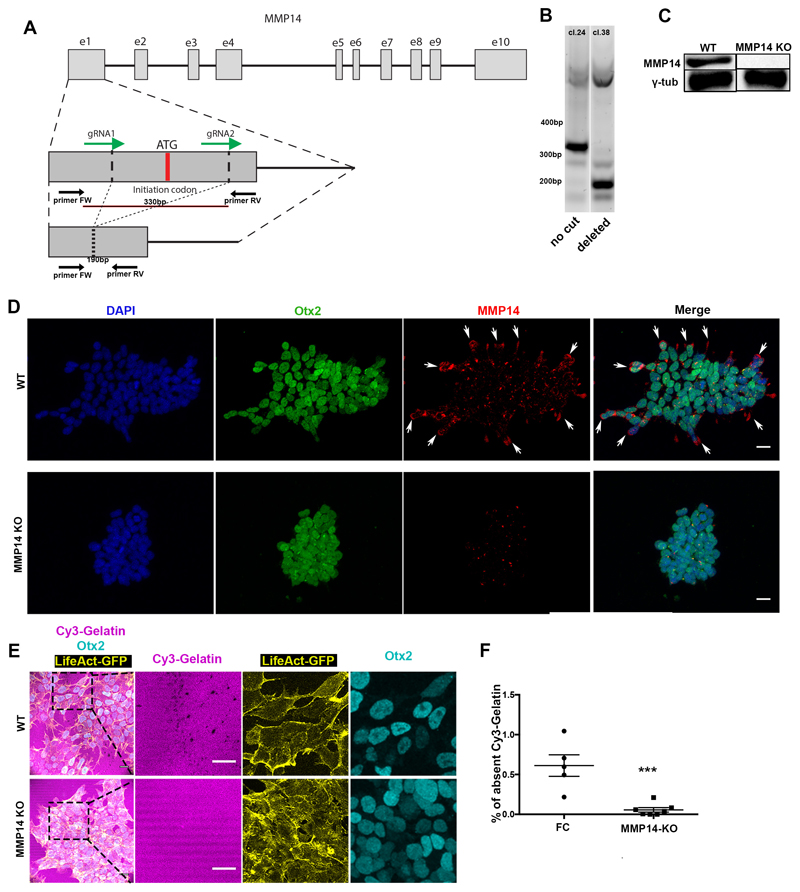
Extended Data Figure 6. MMP14 is necessary for basement membrane remodelling in ESCs.
A) CRISPR-Cas9 exon 1 ATG initiation codon deletion strategy. gRNAs designed to flank a 330bp region which includes the ATG initiation codon. Genotyping primers positions shown as arrows below the exon region. B) Genotyping results of two clones in which in one the exon remained uncut (clone 24) and in the other (clone 38) it was successfully cut. C) Western blot for MMP14 in clone 38 shows successful MMP14 protein abolishment. For gel source data see Supplementary Figure 1. D) Representative examples of control and MMP14 knock-out (KO) Lifeact-GFP ESCs stained for MMP14. MMP14 protein is not detected in the KO cells. 3 independent experiments E) Representative examples of control and MMP14 knock-out (KO) Lifeact-GFP ESCs plated on Cy3-Gelatin. Right panels: Magnified images showing co-localization of control/wildtype ESCs with Cy-3 gelatin perforations. Note the defective extra-cellular matrix remodelling in MMP14-knock-out ESCs. The stripy appearance of gelatin is a result of its manner of plating. 3 independent experiments F) Quantification of (E) of Cy3-Gelatin remodelling based on % of absent fluorescence. Two sided unpaired student’s t-test; ***p<0.001; n=5 regions for FC and 7 regions for MMP14 KO; mean±SEM. FC = feeder cell medium. Scale bars: 20um.