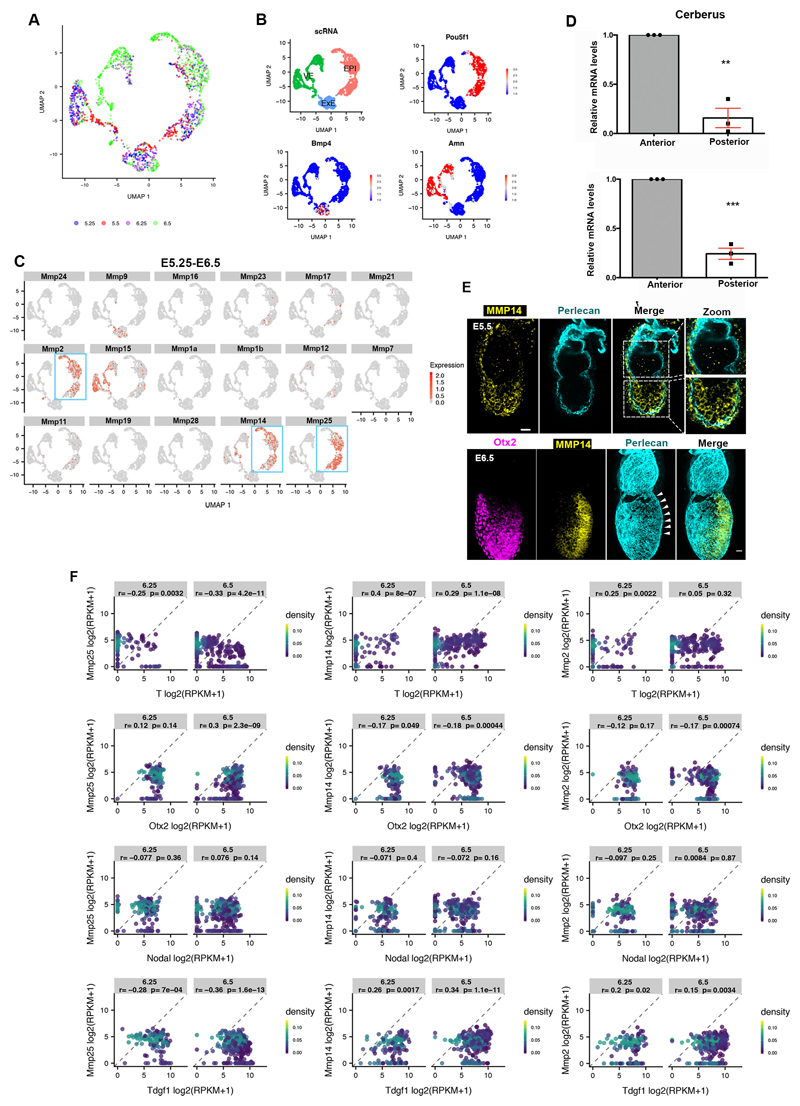
Extended Data Figure 4. MMPs expression profile.
A) UMAP showing distribution of cells by embryonic age. n=1724 cells; E5.25: 331 cells; E5.5: 269 cells; E6.25: 331 cells; E6.5: 803 cells. B) Marker genes identify EPI (Pou5f1), ExE (Bmp4) and VE (Amn) cells in the UMAP. n= 775 EPI cells, 283 ExE cells and 666 VE cells. C) Expression levels of MMP genes on UMAP plot for E5.25-E6.5. Expression values: log(counts). Blue box: EPI. D) mRNA levels of Cerberus and MMP25 at the anterior and posterior side of the embryo. Anterior and posterior identity was defined based on Cerberus expression. MMP25 is highly expressed at the embryo’s anterior side. n=3 embryos. Two sided unpaired student’s t-test. Cerberus:**p=0.001; MMP25 ***p=0.0002; mean±SEM E) E5.5 and E6.5 pre-gastrula embryos stained for MMP14; white arrowheads: prospective primitive streak. n=10 embryos for each stage. Scale bars=20um F) The matrix of RPKM values from GSE109071 was used to calculate gene expression correlation between pairs of genes. Epiblast cells at ages 6.25 and 6.5 (n=527 cells) were extracted from the matrix and Pearson's correlation coefficient (r) with two sided p-value are given for each comparison. The points are coloured by density (kde2d).