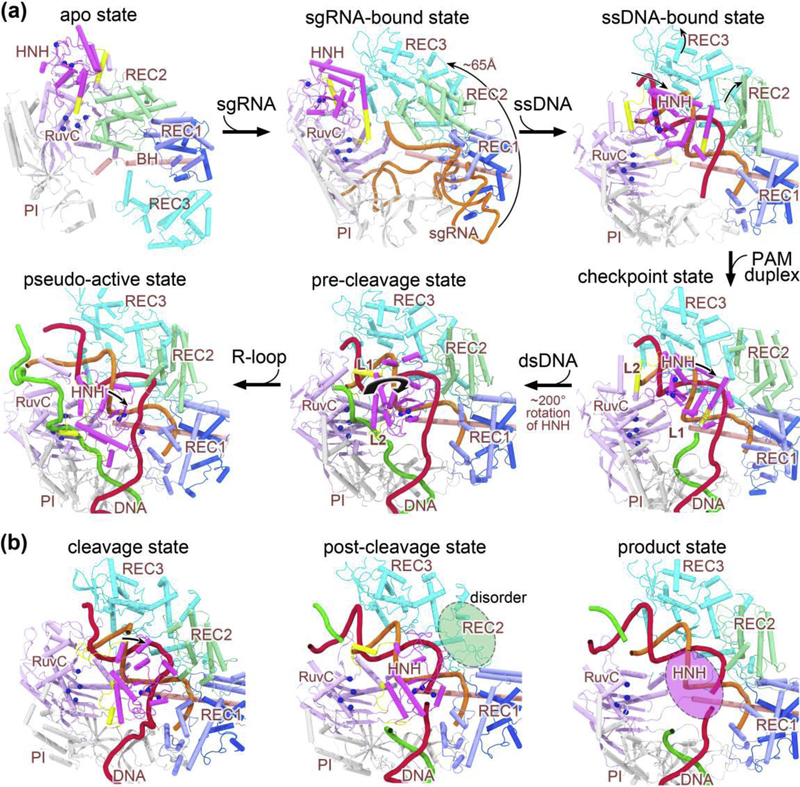
Figure 2.
Stepwise conformational changes in Cas9 leading to cleavage activation. (a) The X-ray and cryo-EM structures of Cas9 reported from years 2014 to 2017 that highlight major conformational changes at each stage of assembly. Along the conformational activation pathway, these structures (shown clockwise) represent the apo state (PDB code: 4CMP), the sgRNA-bound pre-targeting state (PDB code: 4ZT0), the ssDNA-bound state (PDB code: 4OO8), the checkpoint state (PDB code: 4UN3), the pre-cleavage state (PDB code: 5F9R), and the pseudo-active state (PDB code: 5Y36), respectively. (b) The computational model and cryo-EM structures of Cas9 ternary complex presented in 2019 capturing the cleavage state (left), post-cleavage (middle; PDB code: 6O0Y), and product state (right; PDB code: 6O0X). The protein domains are color-coded as in Figure 1a, and the sgRNA, target DNA strand, non-target DNA strand are represented in orange, red, and green cartoon forms, respectively. Note that the split parts of REC1 are colored light and dark blue, respectively. For sgRNA, only the targeting region at its 5’ end is displayed for clarity (except 4ZT0). The alpha-carbon atoms of the active residues in the two nuclease domains (i.e. HNH and RuvC) are depicted as blue spheres. Major domain rearrangements of Cas9 are indicated with curved arrows. Note that the HNH and REC2 domains display alternate disorder transitions (highlighted with translucent ovals) in the post-cleavage and product complexes. Abbreviations: BH, bridge helix; PI, PAM-interacting domain; sgRNA, single guide RNA; ssDNA, single-stranded DNA; dsDNA, double-stranded DNA.