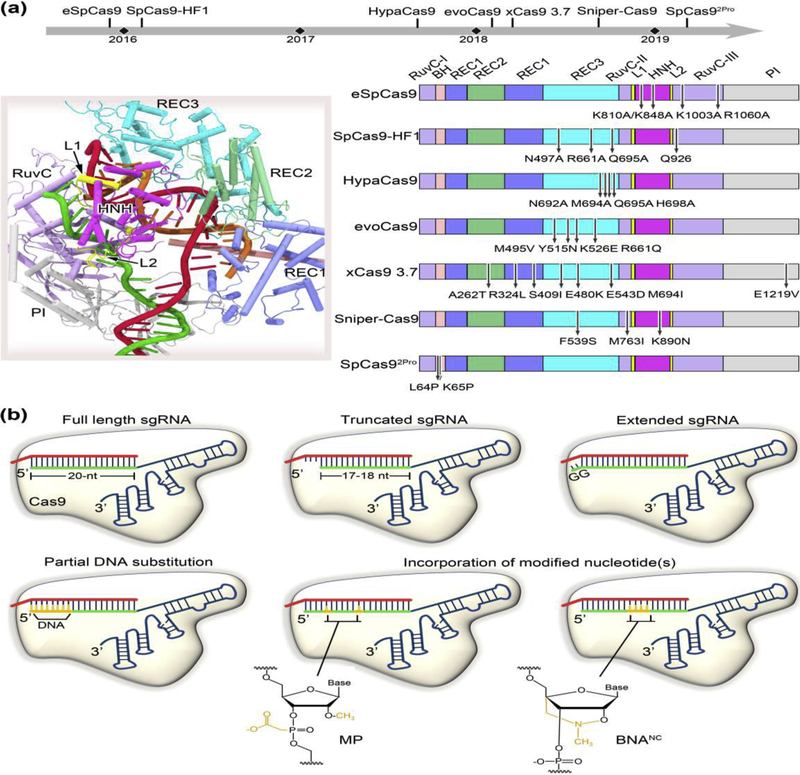
Figure 3.
Allosteric modulation of Cas9 targeting accuracy through protein or sgRNA engineering. (a) Specificity enhanced Cas9 mutants obtained through the strategies of structure-guided rational design or directed evolution. The upper panel shows the timeline of various high-fidelity Cas9 variants reported to date. The PDB structure 5F9R is used for illustration of individual Cas9 domains where the mutations are located (left panel). The mutation positions for each Cas9 variant are marked in the right panel. eSpCas9, SpCas9-HF1, HypaCas9 and SpCas92Pro have been designed by rational approaches, and evoCas9, xCas9 3.7, and Sniper-Cas9 screened through directed evolution. (b) Schematic diagram of sgRNA modifications conferring improved specificity. Most of these modifications occur at the 5’ end of the sgRNA, including truncation of two or three nucleotides, extension of two guanine nucleotides, and partial DNA replacement. The incorporation of chemically modified nucleotides, such as 2’-O-methyl-3’-phosphonoacetate (MP) and bridged nucleic acids (2’,4’-BNANC[N-Me]) at specific locations to the 5’ end or within the central region of the sgRNA guide has also been demonstrated to broadly reduce off-target cleavage.