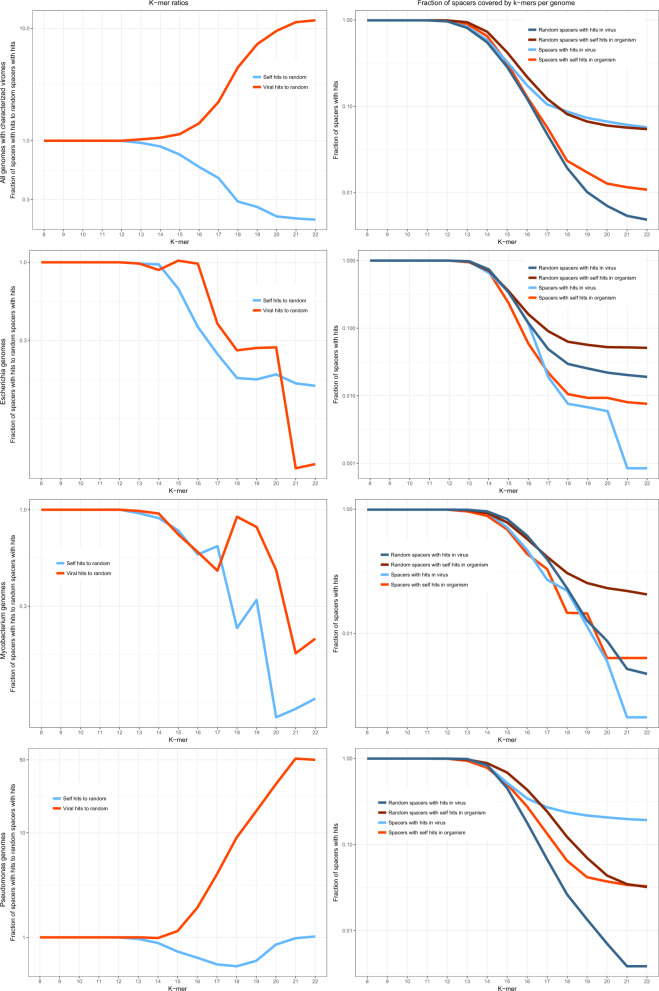
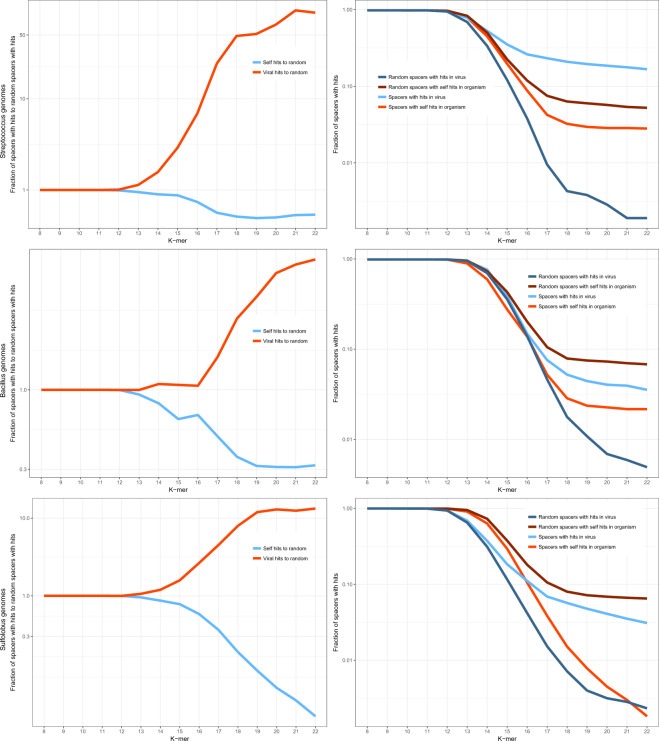
Fig. 3. Comparison of the fractions of k-mer matches for real and mock spacers from prokaryotes with well-characterized viromes.
The top left panel shows a cumulative plot of the fractions of spacers with k-mer matches for 390 host genomes and 2665 viral genomes. Each of the other panels shows the same fractions for the individual bacteria or archaea and the associated viromes. Blue, real spacers matching viral genomes; dark blue, mock spacers matching viral genomes; red, real spacers matching the host genome; dark red, mock spacers matching the host genome. The underlying data are available at ftp://ftp.ncbi.nih.gov/pub/wolf/_suppl/spacers2020/.