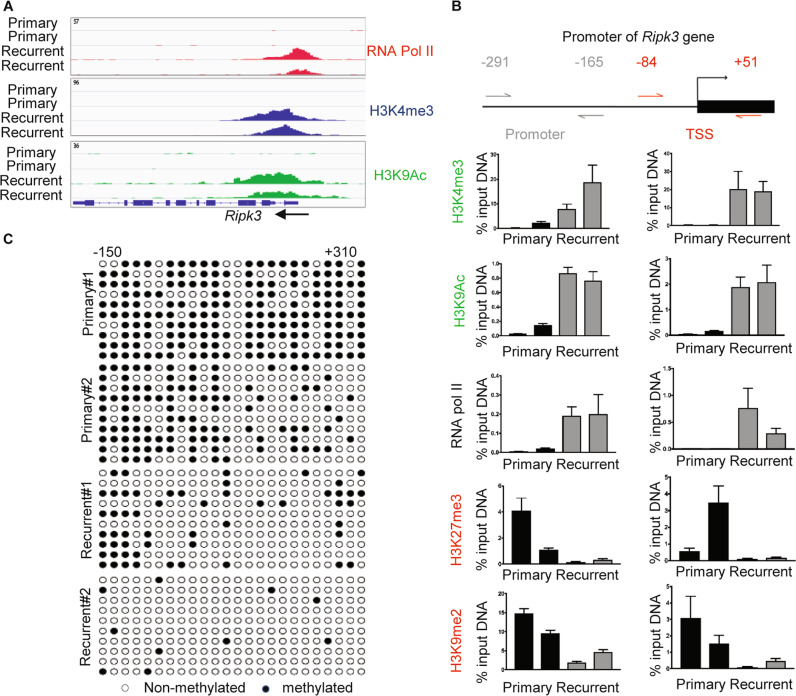
Fig. 2. Epigenetic landscape of the regulatory regions of Ripk3 in the primary versus recurrent tumor cells.
a ChIP-Seq data showed the occupancy of RNA Pol II, H3K4me3, and H3K9ac in regulatory regions of Ripk3 of recurrent tumor cells. b ChIP-qPCR analysis of H3K4me3, H3K9ac, RNA pol II, H3K27me3, and H3K9me2 enrichment at two indicated regions in the promoter of the Ripk3 genes in two primary and two recurrent tumor cell lines. Data are presented as the percentage of input DNA. c The cytosine methylation of CpG dinucleotides (circles) within the Ripk3 promoter and gene body (−150 to +310) for two primary and two recurrent tumor cell lines. Bisulfite-treated DNA was transformed into bacteria and ten replicate colonies were sequenced (rows). Open circles denote unmethylated CpG dinucleotides, while closed circles denoted methylated CpG dinucleotides.