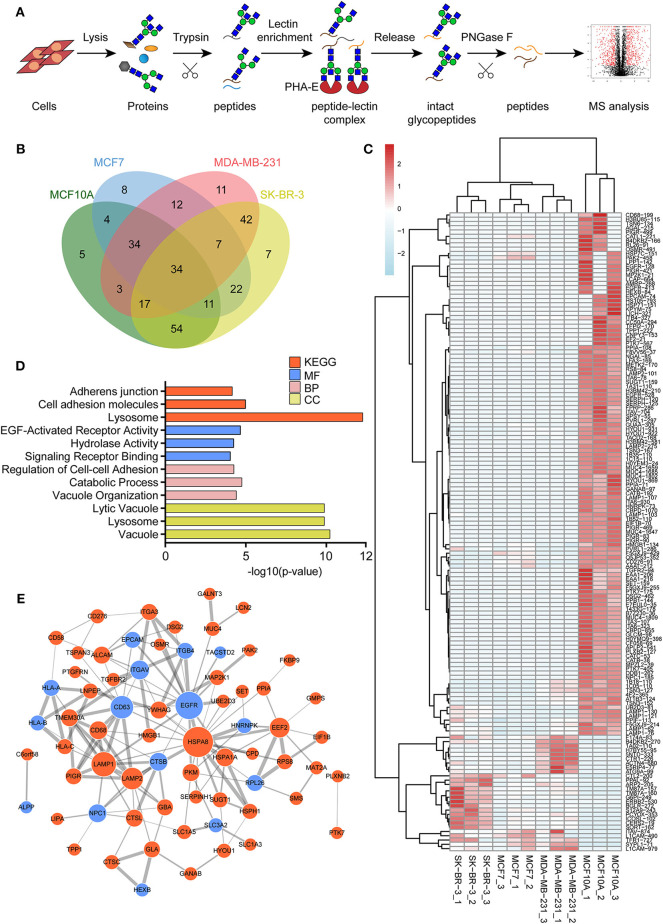
Figure 4.
Identification of glycoproteins bearing bisecting GlcNAc. (A) Workflow of glycoproteomic analysis by combination of PHA-E enrichment and LC-MS/MS. (B) Venn diagram of numbers of identified glycoproteins from MCF10A, MCF7, MDA-MB-231, and SK-BR-3 cells. (C) Heatmap of significantly regulated glycopeptides with bisecting GlcNAc. Each colored cell on the map corresponds to a concentration value of the glycopeptide with identified bisecting GlcNAc glycosylation site. (D) Functional classification of differentially regulated glycoproteins bearing bisecting GlcNAc using Cytoscape plugin BiNGO based on universal GO and KEGG annotation terms. (E) Functional network analysis of differentially regulated glycoproteins using Cytoscape software. Red and blue nodes represented up- and down-regulated glycoproteins. Nodes with more interaction neighbors were displayed in larger size, and edges with larger combined scores were displayed in large size.