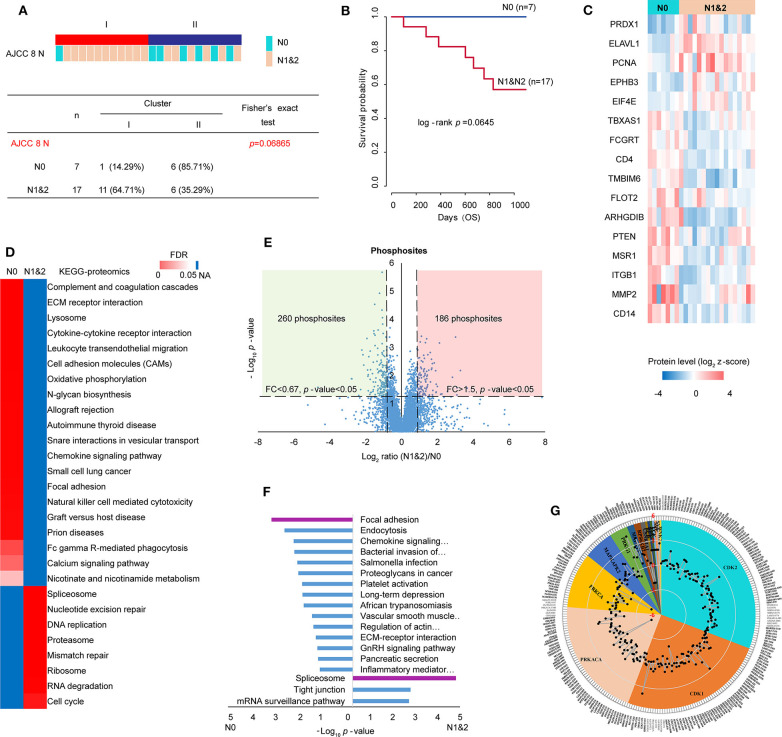
Figure 4.
Analysis of lymph node metastasis (LNM). (A) Relationship of cluster with LNM. Fisher's exact test was adopted for the analysis. (B) The prognosis of patients with or without LNM within 3-year follow-up. (C) 16 differential metastasis-related proteins based on total 355 proteins which were associated with lung cancer metastasis from HCMDB database (fold change> 1.5, p-value < 0.05). (D) GSEA analysis of proteomic data with and without LNM (tumor samples, n = 24): N0 (without LNM, n = 7) and N1&2 (with LNM, n = 17). The heatmap showed the enriched KEGG pathways from MSigDB. The color in heatmap was according to FDR, and the darkest blue represents N/A. (E) Scatter plot depicting the fold change in phosphosites comparing N1&2 with N0. Log2 fold changes were shown on the x-axis and –log10 p-values are shown on the y-axis. The vertical dashed lines indicated fold change > 1.5 and the horizontal dashed line indicated p-value < 0.05 (t-test). (F) DAVID analysis of phosphoproteomic data between N1&2 and N0. The bar chart showed the enriched KEGG pathways of the regulated phosphoproteins between N1&2 and N0 (fold change> 1.5, p-value < 0.05). The highlighted pathways were consistent with those enriched in the protein level. (G) Circular plot represented potential kinases and phosphosites retrieved from the PhosphoSitePlus database. The phosphosites were differentially expressed between N1&2 and N0 with a cutoff of 1.5-fold change. Different colors correspond to various kinases that were predicted as upstream regulators of the phosphosites (p-value < 0.05). The outer circle shows the sites. The radar map shows the fold changes of the sites.