Figure 2. Polymer branching and reducibility can be modulated to control siRNA binding affinity and release kinetics.
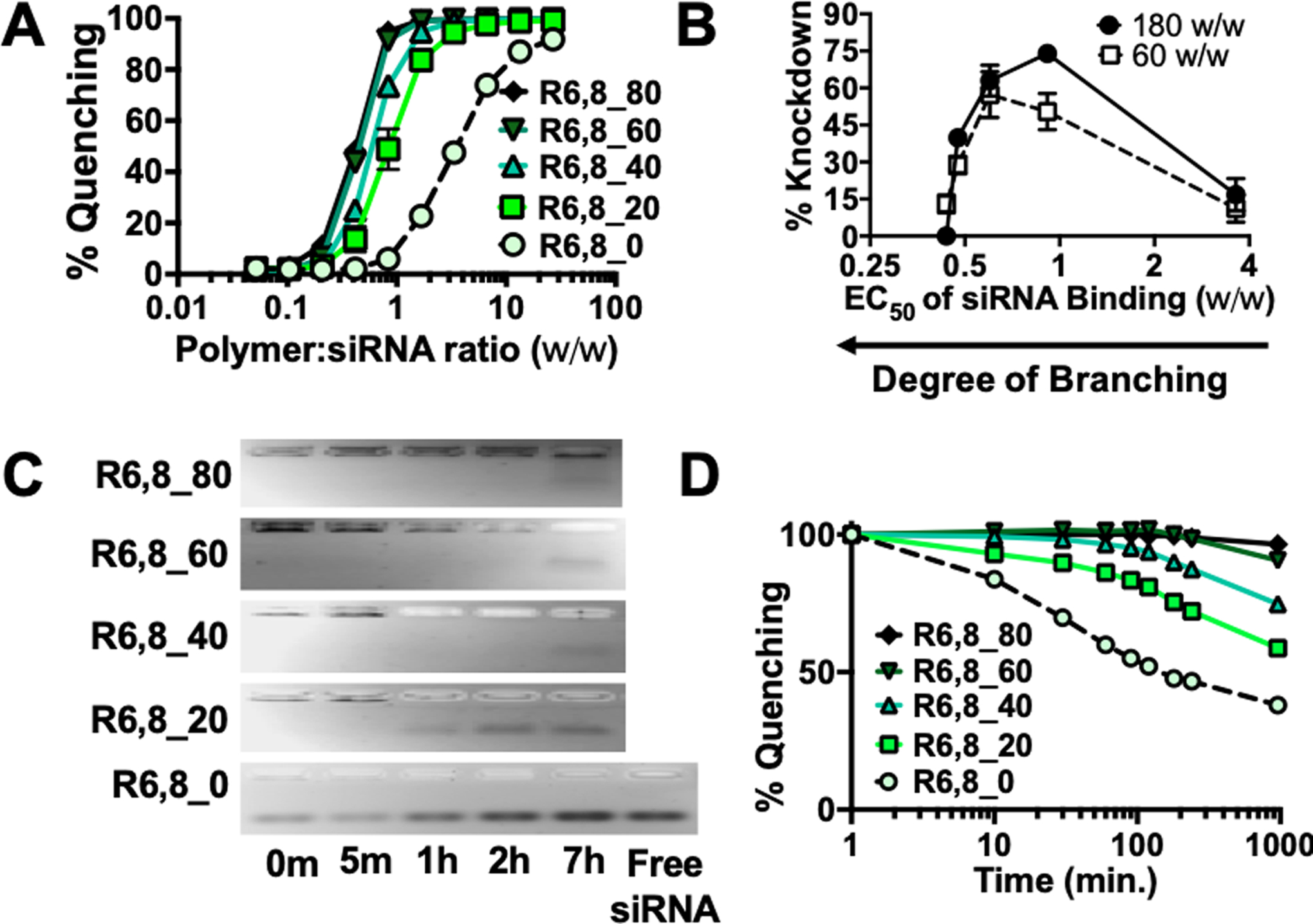
(A) Yo-Pro-1 iodide binding competition assay of R6,8-4-6 polymers to assess siRNA binding affinity. N = 4. (B) % knockdown plotted as a function of polymer EC50 w/w for siRNA binding. N = 4. (C) Gel retardation assays (N = 1) and (D) Yo-Pro binding assay (N = 4) were performed on nanoparticles incubated in 5 mM glutathione to mimic intracellular reducing environments and nucleic acid release slowed as the polymers became more branched with less frequent bio-reducible linkages.
