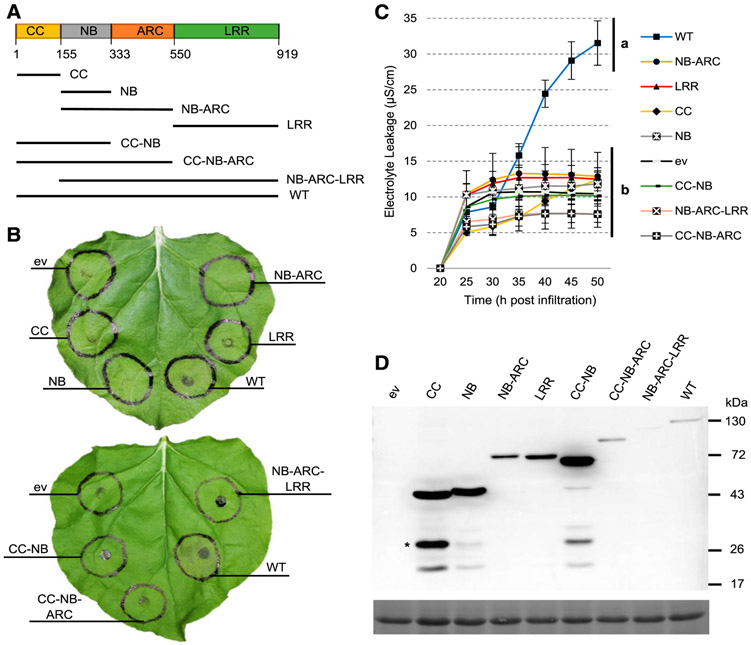
Fig. 2.
Truncation variants of Sr35 are not able to induce cell death. A, Model of Sr35 protein with amino acid numbers used to delineate the boundaries of domains and motifs listed below. CC = coiled-coil, NB = nucleotide binding, NB-ARC = nucleotide binding adaptor, and LRR = leucine-rich repeat. Sr35 wild-type (WT) protein and its truncated variants are shown below the model. B, Macroscopic cell death in Nicotiana benthamiana leaves 48 h postinfiltration (hpi) with Sr35 and its truncated variants. All proteins were C-terminally tagged with green fluorescent protein (GFP); ev refers to empty vector. C, Electrolyte leakage was monitored from 20 to 50 hpi. Error bars correspond to standard error based on four biological replicates per construct. Different letters represent significantly different groups of means, based on Tukey’s honestly significant difference test (α = 0.01) performed at the last timepoint (statistical analyses in Supplementary Table S3). D, GFP-horseradish peroxidase Western blot showing proteins expressed at expected sizes for all constructs. GFP cleavage (*) was observed in CC, NB, and CC-NB constructs. The lower panel is the same blot stained with Ponceau S to reveal bands corresponding to the Rubisco large subunit protein as loading control.