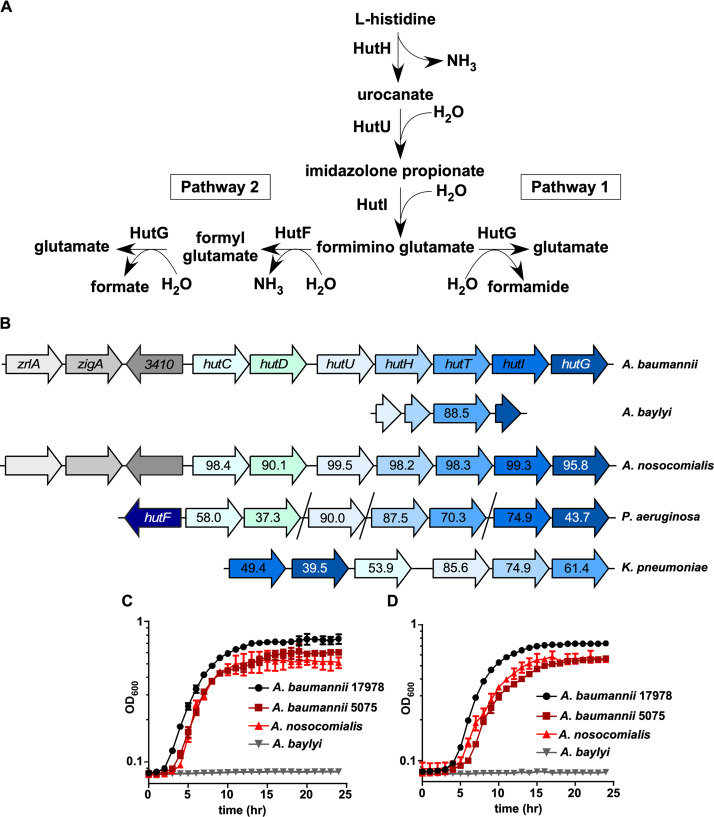
FIG 2.
The histidine utilization system is broadly conserved. (A) Model for the two defined pathways for Hut-mediated histidine catabolism (3). (B) Genomic organization of hut systems in A. baumannii ATCC 17978, A. baylyi ADP1, A. nosocomialis M2, Pseudomonas aeruginosa PA14, and Klebsiella pneumoniae ATCC 43816. Numbers within arrows denote the percent protein identity compared to A. baumannii ATCC 17978. (C) Representative growth curve monitored based on the OD600 over time of A. baumannii 17978, A. baumannii 5075, A. nosocomialis M2, and A. baylyi ADP1 in M9 minimal media with histidine as the sole carbon source. (D) Representative growth curve monitored based on the OD600 over time of A. baumannii 17978, A. baumannii 5075, A. nosocomialis M2, and A. baylyi ADP1 in M9 minimal media with histidine as the sole carbon and nitrogen source.