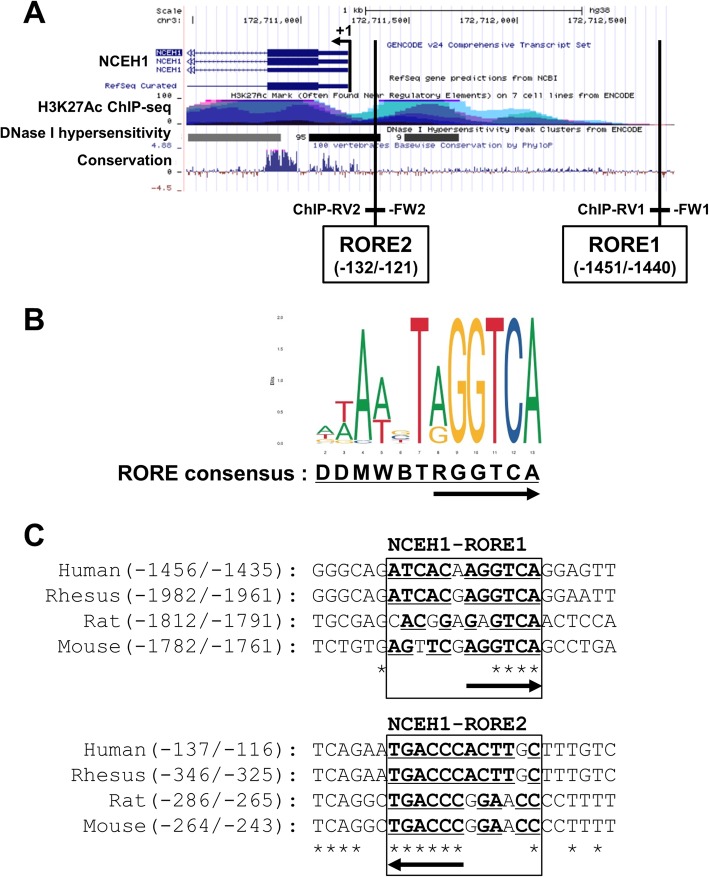
Fig. 1.
RORα response elements in the NCEH1 promoter region. a The UCSC Genome Browser was used to identify distal, conserved, putative RORα response elements (ROREs) in the 5′ region of human NCEH1, which contained the promoter and was located within a DNase I hypersensitivity site. The positions of putative ROREs (RORE1 and RORE2) and transcription start site (TSS, + 1 to position) are indicated by vertical lines. Chromatin immunoprecipitation (ChIP) assays were performed to amplify the ROREs using ChIP-FW1 and ChIP-RV1 or ChIP-FW2 and ChIP-RV2 primer sets. Luciferase reporter constructs of the human NCEH1 promoter region were amplified by PCR using Promoter-FW (− 1689) and Promoter-RV (+ 128) primers. b The JASPAR tool was used to identify RORE consensus sequences (i.e. DDMWBTRGGTCA). RORE half-sites were found to be highly conserved regions, as indicated by arrows. c Schematic representation of putative ROREs in NCEH1 of various species aligned using ClustalW programs. The human sequence shown includes bases − 1456 to − 1435 as NCEH1-RORE1 and − 137 to − 116 as NCEH1-RORE2 in the putative RORE region, which was tested using electrophoretic mobility shift assays (EMSAs) and reporter experiments. The nucleic acid sequence is shown, with RORE consensus sequences underlined and conserved sequences marked with asterisks in mammals. The direction of the RORE half-site is indicated by arrows