Fig. 2.
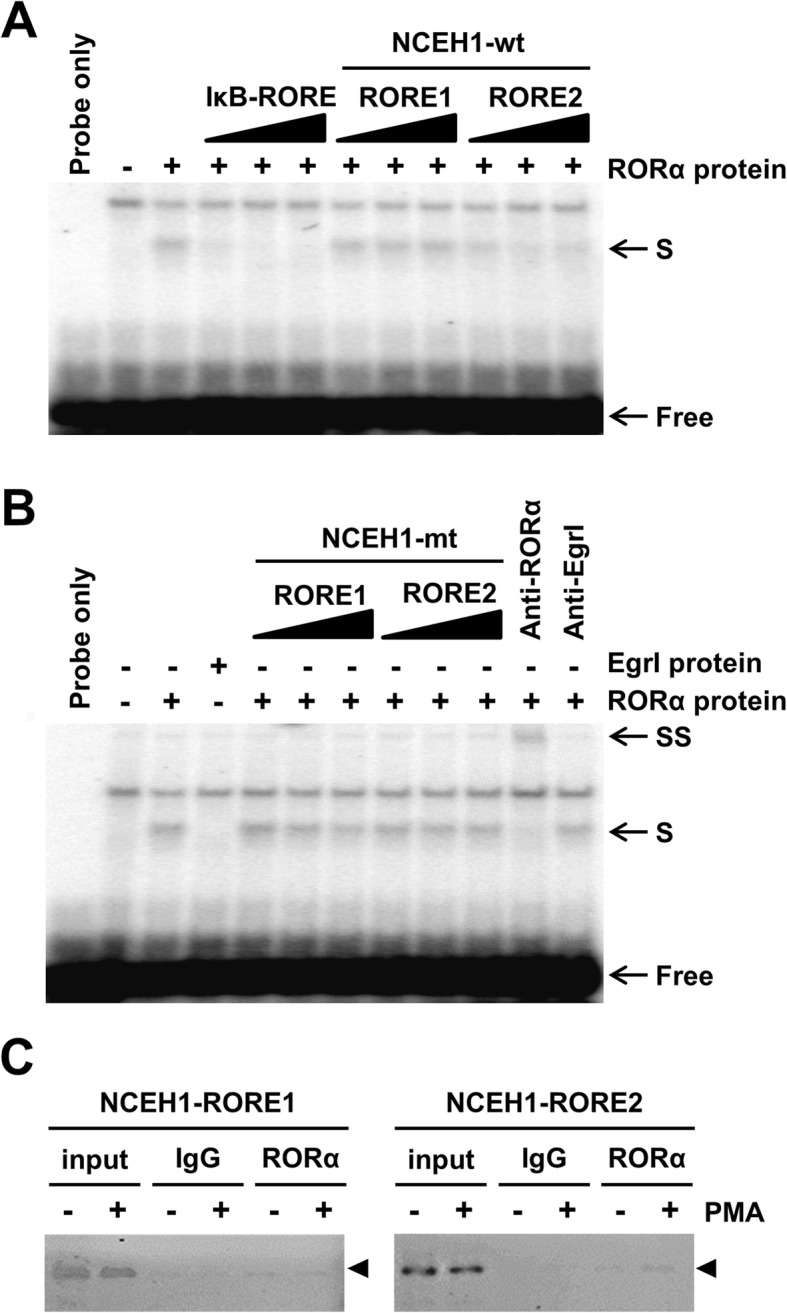
RORα binds a putative response element in NCEH1. a Competition of unlabeled duplexes with the labelled IκB RORE probe for binding to in vitro-translated RORα proteins. The IκB probe contained the known RORα response element (RORE) for NF-κB inhibitor α. Reactions containing RORα proteins were carried out in the absence or presence of a 5-, 10-, and 20-fold molar excess of unlabeled duplexes as competitive probes (IκB-RORE, NCEH1-RORE1, and NCEH1-RORE2 [wild-type]). Shifted binding is indicated by S-arrows. The positions of free probes are indicated by free arrows. b Electrophoretic mobility shift assays (EMSAs) were used to test the ability of unlabeled mutated probes (5-, 10-, and 20-fold molar excess) to relieve the inhibition of binding of RORα to the putative response element (S-arrows). Anti-RORα antibodies were pre-incubated with RORα protein before adding the labeled probe for the formation of the super-shift band (SS-arrows). Negative control experiments were performed using EgrI protein and antibodies. Lane 1: labelled probe only; lane 2: reaction containing crude products from in vitro translation; lane 3: probe incubated with crude product obtained from in vitro translation in the absence of the RORα expression vector. c Chromatin immunoprecipitation (ChIP) assays were performed using chromatin isolated from human monocytes and differentiated macrophages treated with 100 nM phorbol 12-myristate 13-acetate (PMA) for 24 h. Crosslinked cell lysates were immunoprecipitated with rabbit IgG (IgG) or polyclonal anti-RORα-specific antibodies (RORα). DNA precipitates were isolated and then subjected to PCR using primer pairs covering either RORE1 or RORE2 of the NCEH1 promoter region. Control PCR was performed with non-immunoprecipitated genomic DNA (input)
