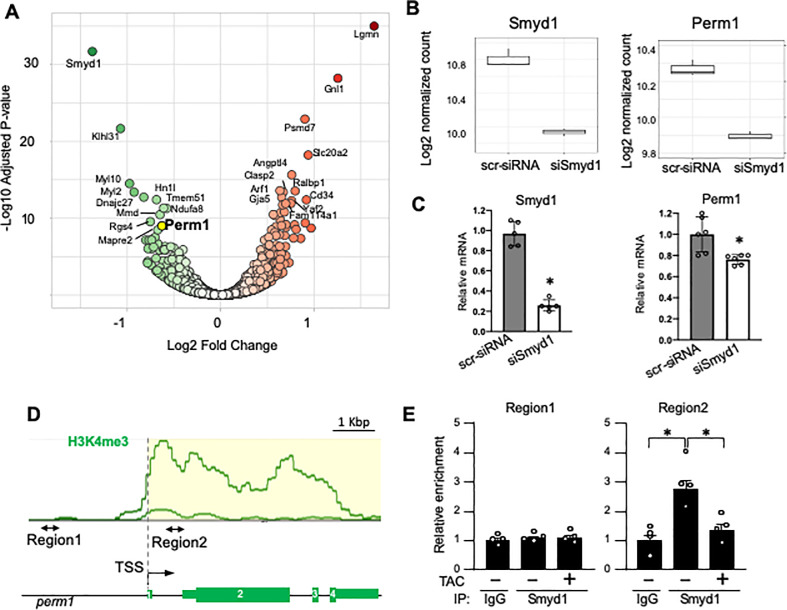
Fig 1. Perm1 is a downstream target of Smyd1.
(A-B) RNA-seq of siSmyd1 cardiomyocytes. A volcano plot shows significantly downregulated (green) and upregulated (red) genes. Perm1, indicated by a yellow circle, is one of the genes most significantly downregulated by siSmyd1 (Panel A). Box plots show the decreased mRNA levels of Perm1 in cardiomyocytes in which Smyd1 expression was significantly reduced by siSmyd1 as compared with control cells that were treated with scrambled-siRNA (scr-siRNA) (n = 4/group, Panel B). (C) qRT-PCR showing a significant reduction in Perm1 in cardiomyocytes in which Smyd1 was knocked-down by siRNA (n = 6/group). Statistics were performed using the t-test with 2 tails. *p<0.05. (D) Previous ChIP-seq studies have established enrichment of H3K4me3 within the promoter region of the Perm1 locus (WashU EpiGenome Database), which was targeted for qPCR reaction (“Region 2”). As a negative control, the ~2k bp upstream of the transcription start site (TSS) was targeted (“Region 1”), where small H3K4me3 peaks were observed. (E) ChIP-PCR showing no significant interaction of Smyd1 with Region 1 (left), while Smyd1 does bind to Region 2 in the hearts of WT mice. This interaction was significantly reduced in the TAC heart (TAC +) (right) (n = 4 in sham, n = 4 in TAC). Statistics were performed using the t-test with 2 tails. *: p<0.05. Error bars are ±SEM.