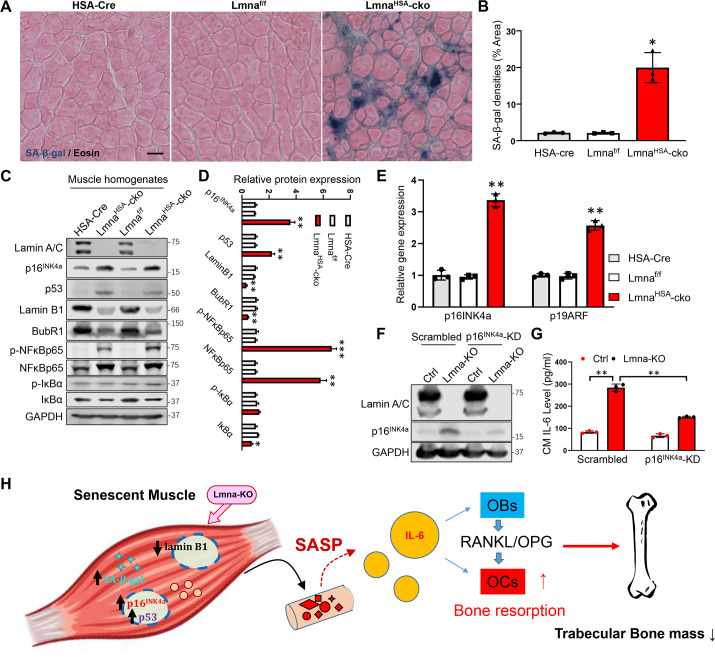
Fig 8. Increased cellular senescence in Lmna-KO muscles and the requirement of p16INK4a for the increase of IL-6 expression in Lmna-KO muscle cells.
(A) SA-β-gal staining counterstained with eosin of gastrocnemius cross-section from 3-mo HSA-Cre, Lmnaf/f, and LmnaHSA-cko mice. Scale bar, 20 μm. (B) Quantification of SA-β-gal densities (mean ± SD; n = 3). *P < 0.05, significant difference. (C) Western blot analysis of indicated protein expression in muscles from mice with indicated genotypes (at 1 mo). GAPDH was used as a loading Ctrl. (D) Quantification analysis (mean ± SD; n = 3). *P < 0.05, **P < 0.01, ***P < 0.001. (E) Real-time PCR analysis of p16INK4a and p19ARF expression in muscles from mice with indicated genotypes (at 1 mo) (mean ± SD; n = 3). **P < 0.01, significant difference. (F) Western blot analysis of p16INK4a, which was reduced in p16INK4a-KD muscle cells. (G) ELISA analysis of IL-6 level in CM of Ctrl, Lmna-KO, p16INK4a-KD, and Lmna-KO; p16INK4a-KD C2C12 muscle cells (mean ± SD; n = 3). **P < 0.01, significant difference. (H) Illustration of a working model. Lmna loss in skeletal muscles results in increased cellular senescence and p16INK4a, which increases the expression and secretion of SASP factors, including IL-6, thus up-regulating RANKL expression in OBs and promoting osteoclastogenesis and trabecular bone loss. The underlying data for this figure can be found in S1 Data. BubR1, Bub1-related kinase; cko, conditional knockout; CM, conditioned medium; Cre, cyclization recombination enzyme; Ctrl, control; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; HSA, human alpha-skeletal actin; IκBα, inhibitor of nuclear factor kappa B; IL, interleukin; KD, knockdown; KO, knockout; Lmna, lamin A/C gene; Lmnaf/f, floxed Lmna mice; LmnaHSA-cko, skeletal muscle–specific Lmna-cko mice; mo, months old; NF-κB, nuclear factor kappa-light-chain-enhancer of activated B cells; OB, osteoblast; OC, osteoclast; OPG, osteoprotegerin; p19ARF, the alternate reading frame tumor-suppressor protein; RANKL, receptor activator of NF-κB ligand; SA-β-gal, senescence-associated beta-galactosidase; SASP, senescence-associated secretory phenotype.