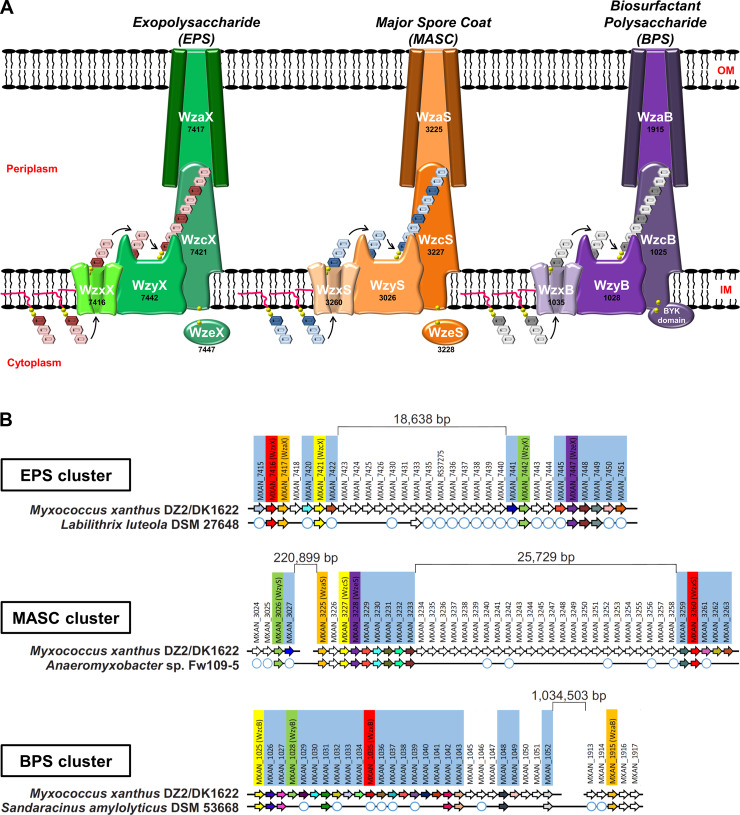
Fig 1. Wzx/Wzy-dependent polysaccharide biosynthesis pathways encoded by M. xanthus.
(A) Schematic representation of the 3 Wzx/Wzy-dependent polysaccharide-assembly pathways in M. xanthus DZ2/DK1622. The genomic “MXAN” locus tag identifier for each respective gene (black) has been indicated below the specific protein name (white). (B) Gene conservation and synteny diagrams for EPS, MASC, and BPS clusters in M. xanthus DZ2/DK1622 compared to the evolutionarily closest genome containing a contiguous cluster (see S1 Fig). Locus tags highlighted by pale blue boxes correspond to genes such as enzymes involved in monosaccharide synthesis, modification, or incorporation into precursor repeat units of the respective polymer. White circles depict the presence of a homologous gene encoded elsewhere in the chromosome (but not syntenic with the remainder of the EPS/MASC/BPS biosynthesis cluster). BPS, biosurfactant polysaccharide; BYK, bacterial tyrosine autokinase; EPS, exopolysaccharide; IM, inner membrane; MASC, major spore coat; OM, outer membrane.