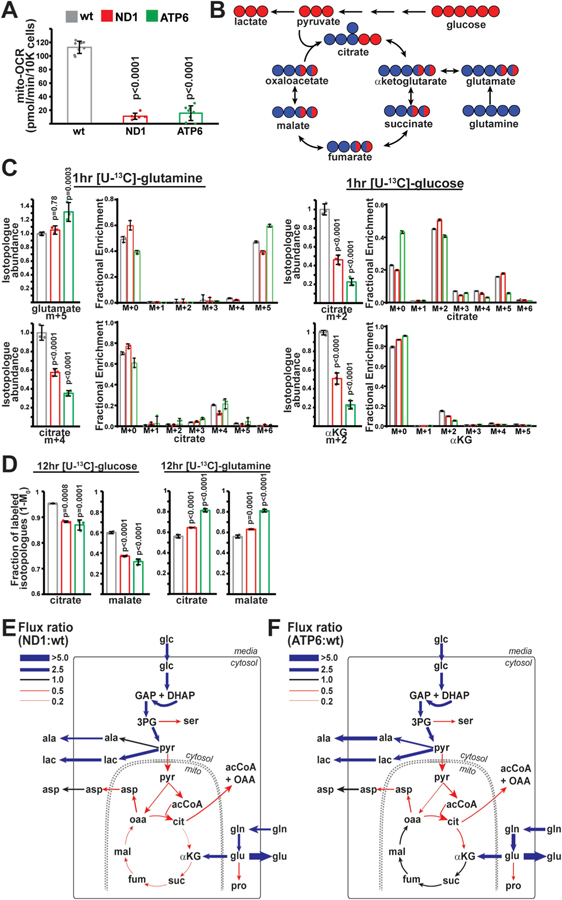
Fig. 1. Metabolic flux analysis of ND1 and ATP6 mutant cell lines.
(A) Oxygen consumption rates in wild-type (wt; n = 8), ND1 (n = 6) and ATP6 (n = 8) mutant cell lines. (B) Schematic of carbon incorporation from glucose (red) or glutamine (blue) into TCA cycle metabolites. (C) Relative isotopologue abundances of the indicated metabolites following 1 h pulses with labeled glucose or glutamine. Abundances (n = 3 for each group) were normalized to total ion count. Same color scheme as (A). (D) Fractional contribution (1-M0) of glucose or glutamine to the indicated metabolites after steady-state labeling with glucose or glutamine. n = 3 for each group. Same color scheme as (A). (E) Flux ratios for individual enzymatic steps between ND1 and wt cells following metabolic flux analysis. Thickness of the line indicates the magnitude of the ratio. Blue represents fluxes upregulated in ND1 cells; red represents fluxes downregulated in ND1 cells (relative to wt). ala, alanine; glc, glucose; lac, lactate; pyr, pyruvate; acCoA, acetyl-CoA; cit, citrate; αKG, α-ketoglutarate; glu, glutamate; gln, glutamine; suc, succinate; fum, fumarate; mal, malate; oaa, oxaloacetate; asp, aspartate; ser, serine; GAP; glyceraldehyde 3-phosphate; DHAP, dihydroxyacetone phosphate; 3 PG, 3-phosphoglycerate; pro; proline. (F) Same as E, but comparing ATP6 and wt cells. Representative data from a single experiment is shown, and all experiments were repeated at least in triplicate. Error bars represent standard deviations from mean of independent samples. Adjusted p-values calculated by 1-way ANOVA, followed by Tukey’s test for multiple comparisons. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)