Figure 5.
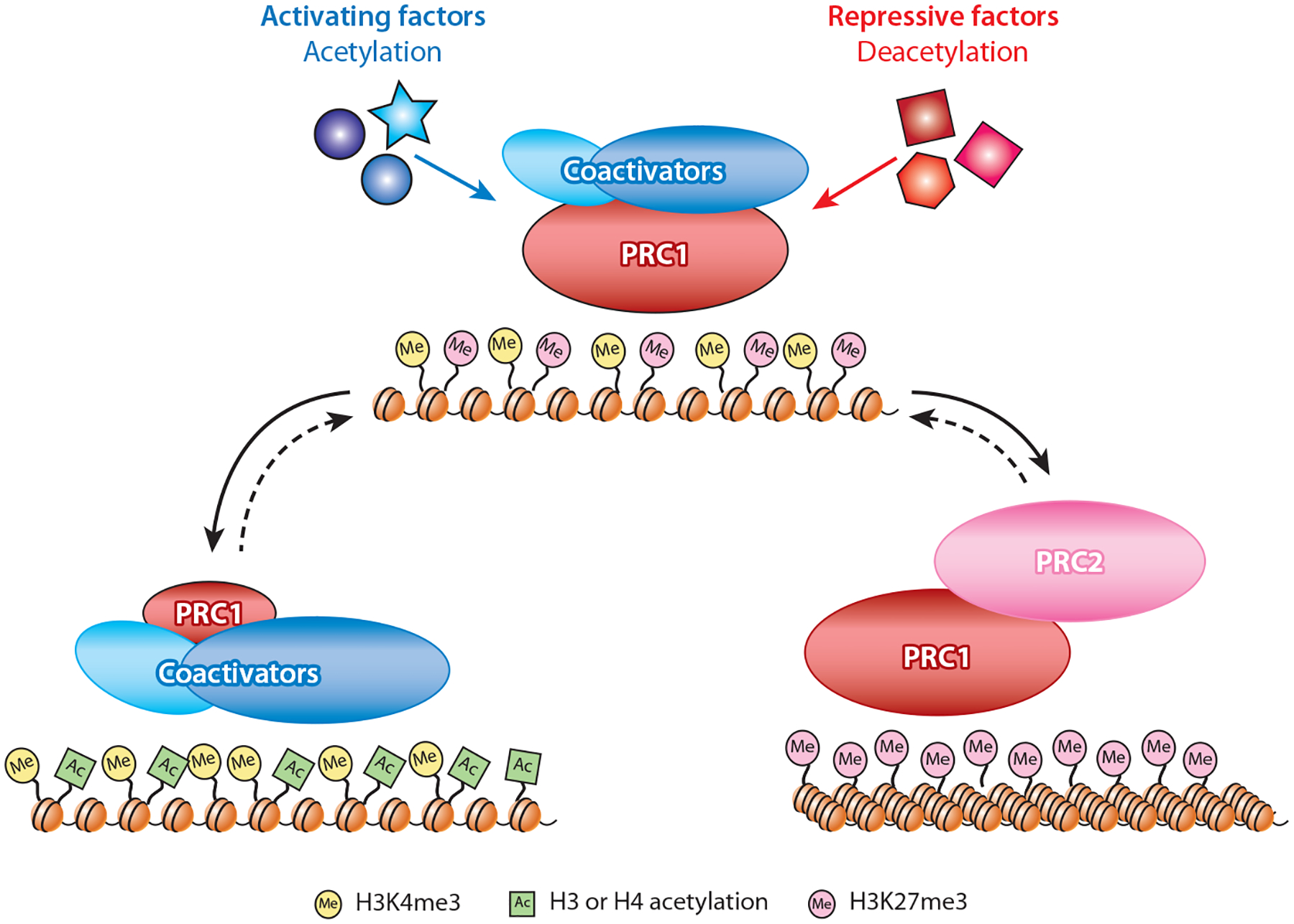
The bivalent master switch model. During embryonic development in Drosophila, PRC1 and specific coactivator proteins are proposed to form bivalent protein complexes on transcriptionally poised genes. The coactivator module includes a histone acetyltransferase that can both catalyze and recognize acetylation marks, whereas a separate subunit can bind the H3K4me3 chromatin mark at active or bivalent promoters. In contrast, PRC1/PRC2 enrichment is reinforced by the H3K27me3 silencing mark and unacetylated nucleosomes. The choice between a transcriptionally active or silent state may be triggered by combinations of specific transcription factors that alter the acetylation state of the local chromatin environment, favoring increased association of coactivators or PRC2, respectively. If some level of co-occupancy is maintained, the resulting transcriptional states may be reversible (dotted lines) by changes in critical thresholds of competing transcription factors during subsequent differentiation. Although this model rests largely on protein interactions observed in Drosophila, it is compatible with the responsive model for PcG targeting in mammalian cells (95). In an extension of that model, mammalian PcG and TrxG proteins may compete for interaction at CpG islands and be influenced by local acetylation, without necessarily interacting physically as observed in Drosophila embryos. Abbreviations: PcG, Polycomb Group; PRC1 and PRC2, Polycomb Repressive Complexes; TrxG, Trithorax Group.
